Recent advancements in technology have now made it feasible for scientific researchers to be able to collect large amounts of biological data using a huge variety of methods, thus allowing scientists to make insightful conclusions. However, these datasets are so massive that they can’t be analyzed manually. Instead, various computational tools are used to make data analysis more efficient. A new tool, AutoBA, has now been introduced that can be used to automate bioinformatics analysis.
High-throughput techniques have allowed scientists to extract data on an unprecedented scale. Bioinformatics aims to solve biological problems through interdisciplinary methods that integrate biological data, data science, artificial intelligence, and machine learning, among other fields. The creation of tools like AlphaFold, which revolutionized protein prediction technology, and the ever-growing array of tools and software packages that are customized to be able to clean, assess, analyze, and visualize biological data are ample proof of its utility and growing popularity.
The Use of Artificial Intelligence and Large Language Models in Bioinformatics
Over the past year, the release of ChatGPT and other AI generation tools has generated much excitement among scientists and the public. Large Language Models (LLMs), in particular, show a lot of promise as tools that can significantly enhance research workflows and make them more efficient. This may have applications in many fields, including drug discovery, systems biology, and disease diagnosis.
While existing LLM tools like ChatGPT and AutoGPT are capable of complex tasks like code generation and data analysis to achieve objectives that the user has defined, these tools aren’t suitable for the complex and intricate tasks that bioinformatics projects normally require. For example, bulk RNA-seq necessitates the performance of a series of complex steps, starting at quality control and progressing to adapter trimming, data cleaning, transcriptome or genome alignment, and more. Another technique, like ChIP-seq, will require different steps. Hence, different methods will require a comprehensive understanding of the relevant technique as well as the programming and data analysis skills necessary to draw conclusions from the data.
One of the complications that bioinformatics analysis poses to conventional tools is the fact that data is obtained from diverse sources, like RNA-seq, ATAC-seq, and WES, among others. Different kinds of data necessitate the use of multiple tools. The burden of researching available options, selecting a suitable tool, and configuring it to their needs falls entirely on researchers, resulting in the loss of valuable time and reduced efficiency.
Online platforms that conduct bioinformatics analysis are gaining popularity, but their nature lends itself to privacy and data security concerns. Since the data being used is biological, it is highly probable that it contains sensitive, private information that may be exposed to malicious elements, compromising privacy and leading to a lack of trust among the public if such incidents become common.
Another common concern is the lack of reproducibility and consistency across different pipelines, resulting in confusion and errors. The process of running such analyses digitally can often be tedious and time-consuming even for experts in the field and can be even more difficult for scientists who do not have the required skills in analysis and dry lab techniques.
Introducing AutoBA – an LLM Tailored for Bioinformatics
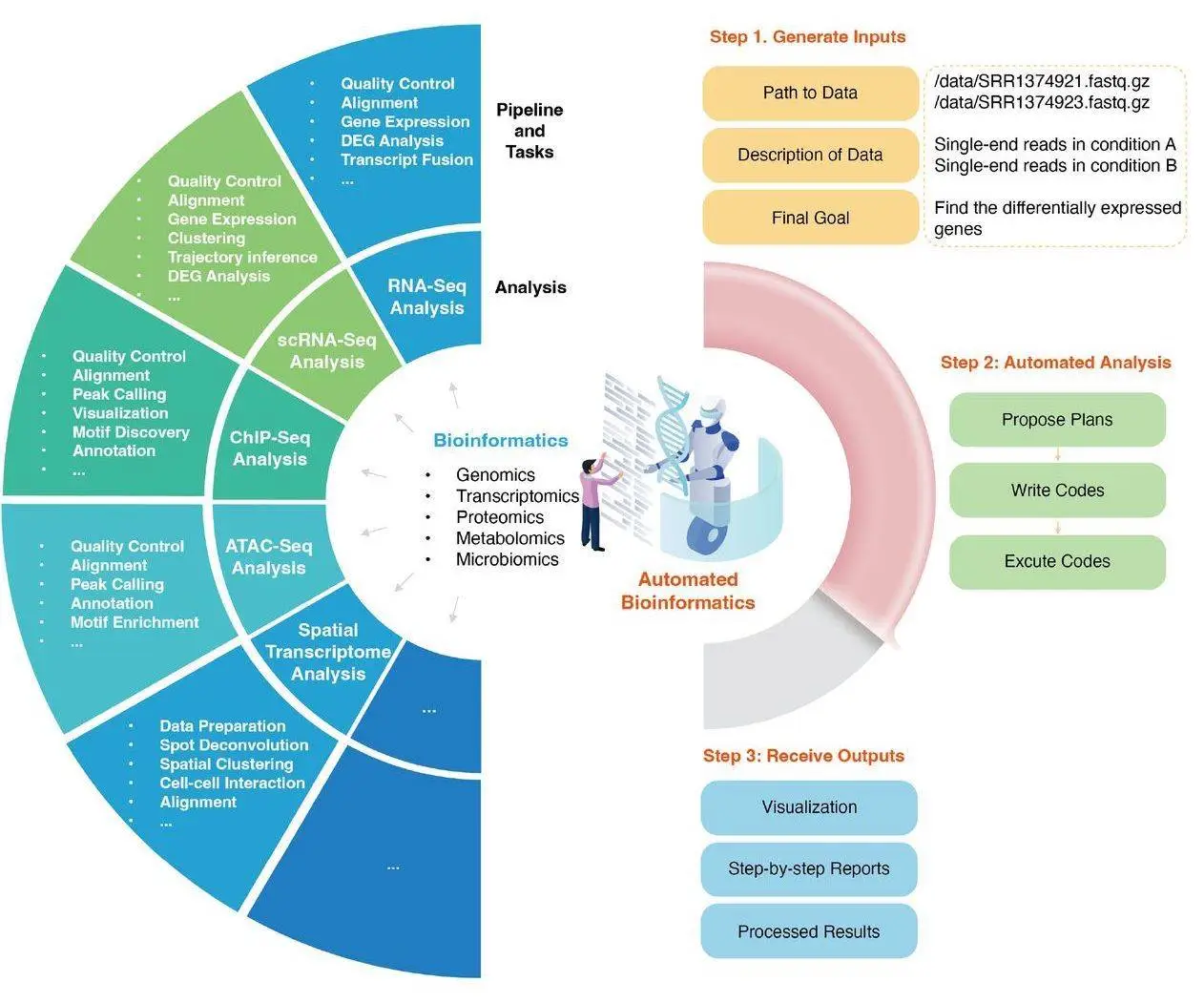
To deal with these issues, a new tool has been developed that has been tailored to the needs of bioinformatics analysts. AutoBA, or Auto Bioinformatics Analysis, is an autonomous software that utilizes Artificial Intelligence to optimize users’ workflow. Only three inputs are required from the user: the data path, description, and objective. It can independently create plans for data analysis, write and execute codes, as well as perform analysis on the data given to it. Researchers do not need to install and familiarise themselves with different tools. AutoBA analyses the data provided by the user, generates a suitable plan, creates code, and analyses the results.
What differentiates AutoBA from other similar data analysis tools is its use of two separate phases, the planning phase, and the execution phase, as part of its analysis process. In the first phase, AutoBA creates a step-by-step blueprint for the analysis task. Subgoals and additional details are included for every stage. In the second phase, AutoBA systematically executes the steps that it previously outlined in the planning phase by performing environment configuration, software installation, and generating and executing code. Since these are considered two separate phases, AutoBA requires a different prompt for each to optimize the performance of analysis tasks. AutoBA also provides ways for users to prevent using certain software by utilizing a blacklist.
Its efficiency has been increased through the incorporation of an ingenious mechanism, where the software can call back to past analyses performed in order to inform its code generation. Outcomes for all steps are logged and become the basis for later projects. This allows AutoBA to save time and reduce redundancy.
AutoBA was tested to check its ability to tackle many bioinformatics tasks, including RNA-seq, single-cell RNA-seq, ChIP-Seq data analysis, etc. AutoBA also tailors itself to user requirements, allowing it to adapt to various inputs and objectives in a way that other tools cannot.
The code and the procedure that AutoBA utilized to generate results were then subjected to scrutiny by human subject-matter experts. This examination demonstrated that the program created precise code for each step and consistently produced dependable results.
Conclusion
AutoBA is a recently developed software; hence, it has not been adequately tested for its proficiency in performing different bioinformatics tasks. Also, as an LLM, it can only give information based on its training. Due to this, techniques developed after AutoBA’s creation and development will likely be complex for it to perform, as it will not have a template to model its analysis after. However, its adaptable nature and efficiency may result in it becoming a valuable tool for research.
Article Source: Reference Paper | AutoBA is available on GitHub
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.