Over the past few years, a large number of single-cell genomics datasets that profile a range of tissues in different stages of development have been created. These resources have been incorporated in the creation of a standard reference map for human bodies by consortia like the Human Cell Atlas. However, different labs may have different definitions of the distinctions between various cell types, hence producing inconsistencies in classification. A new tool, CellHint, developed by the Wellcome Sanger Institute, the University of Cambridge, EMBL’s European Bioinformatics Institute (EMBL-EBI), and their collaborators, aims to remedy this problem.
The ongoing effort to develop a Cell Ontology database aims to rectify this by providing consistent labels and unifying the definition of cell types across communities. Other contributions to this effort include the Human BioMolecular Atlas Program by the NIH and the Cell Annotation Platform. However, beyond mere nomenclature, the standardization of cell types is a task that has many obstacles – discrepancies in annotation resolution and variance in cell quality, among others.
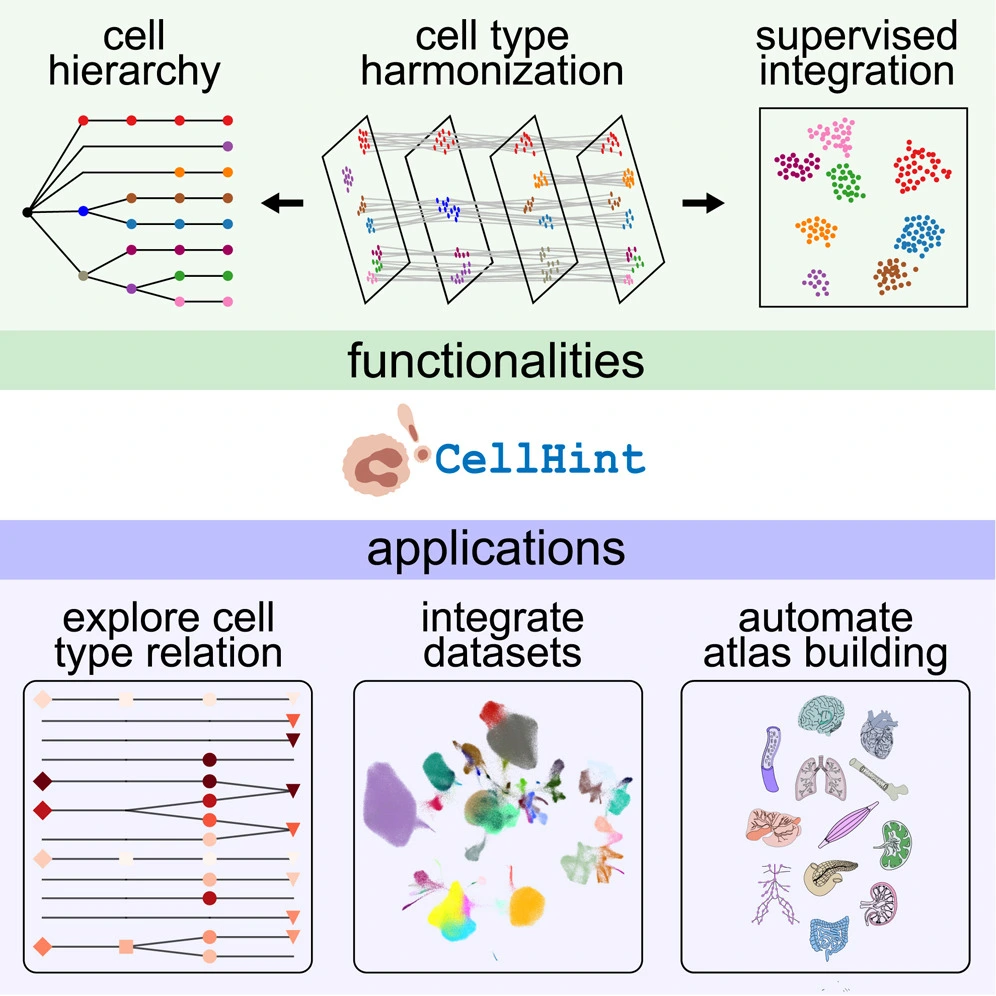
Different methods were developed to match cell types across different datasets. However, they are limited due to the fact that they rely primarily on pairwise dataset alignment – multiple datasets cannot be aligned simultaneously due to the arduousness of the task and the challenges it poses with regard to consistent visualization and information integration. It is further limited due to its tendency to neglect relationships between various cell types when not possessing a prebuilt hierarchy.
Further, recent efforts for data integration have largely focused on the correction of batch effects across different datasets. Information regarding various cell types cannot be fully integrated due to inconsistencies in nomenclature. Once harmonized successfully, it is possible to supervise data integration efforts, taking into account the information provided in annotations. Though methods like scANVI and scGen include such information in their pipelines, further control over the process is required, along with the ability to integrate cell-type relationships into it.
In order to remedy this, a new automated workflow has been developed for the purpose of cell-type harmonization – a new tool, CellHint, was developed to assess cell-cell similarities and harmonize annotations, thus resulting in the alignment of multiple datasets. As such, the semantic relationships between types of cells are defined, and their hierarchies are captured. This information is leveraged as a guide for downstream data integration. The pipeline was applied to 49 datasets, showcasing its utility and efficacy in harmonizing data while providing a useful tool to researchers.
CellHint is capable of inferring relationships between cell types across datasets. It does so by making predictions regarding intracellular distance and providing summaries of cell-type alignments. The algorithm doesn’t rely on any pre-existing assumptions about distributions of gene expression; the tool is capable of producing batch-insensitive dissimilarity measures, thus facilitating a robust analysis across different datasets.
Five curated datasets were selected to validate the cell harmonization pipeline, which consisted of different cell type compositions, tissue coverages, and developmental stages, representing a scenario similar to what the tool would be expected to see in a practical setting. Significant batch effects were observed when visualizing cells onto the UMAP (uniform manifold approximation and projection). CellHint was able to predict transcriptomic distances among the cell types and revealed finer underlying structures while resolving complex alignments.
Many cell types were harmonized hierarchically, resulting in reclassifications of certain subtypes. The harmonization module can also detect homogenous subtypes that cannot be easily separated through cluster-based approaches. CellHint also discovered new cell types that had been previously overlooked.
CellHint was also tested on its applicability to datasets other than single-cell transcriptomes. Five single-nucleus and single-cell datasets were collected, which resulted in the display of 42 cell types at the high hierarchy level and 55 cell types at the low hierarchy level, and the vast majority of these echoed transcriptome-based cell type alignments.
Cell diversity is often known to increase significantly in the presence of diseased conditions as similar cell types undergo molecular reorganization and novel cell types emerge. In order to test the utility of CellHint in such a scenario, the tool was applied to four datasets. Harmonization graphs allowed researchers to locate various cell types and subtypes that had been overlooked in previous studies and interpret cell states in pathological conditions. Different cell types could be clearly separated into diseased and healthy states, thus facilitating a comprehensive analysis of changes in the cellular transcriptome.
After the reannotation of cells, an integration module is also provided that allows for the data structures to be tuned towards harmonized cell types and assists in the mitigation of effects caused by batch confounders. In order to demonstrate the efficiency of its automated workflow, CellHint was utilized to develop multi-organ reference maps through manually collecting public datasets, automated harmonization and integration, and the curation and finalization of the atlases.
Conclusion
The use of the PCT (predictive clustering tree) algorithm, along with critical optimization steps, results in CellHint possessing great speed, accuracy, and scalability. When its performance was evaluated, it was found that CellHint could successfully redefine and reannotate cell types while incorporating information regarding their relationships. Such an approach resolves the obstacle of dataset order instability and multi-dataset alignment while generating consistent hierarchies among cell types.
The reconstruction of such hierarchies has been a recurrent challenge in the field of cell biology due to the difficulty of segregating cell types. Incorporating manual annotations and integrating the associated information across datasets allows for the development of an approximation of biological hierarchies. Variations across datasets can also be mitigated, facilitating the discovery of cell-type relationships in a batch-insensitive manner. The highly structured architecture of CellHint, in tandem with the multi-organ atlases developed through it, has the potential to aid in the creation of single-cell resources and improve our understanding of cell types across organs and in different pathological conditions.
Article Source: Reference Paper | Reference Article | CellHint (Version: 0.1.1) is available at GitHub
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.













[…] CellHint Revolutionizes Human Cell Atlas: Automatic Cell Type Harmonization Paves the Way for New Di… […]