TBtools, a toolkit used for data analysis and bioinformatics tasks, was released in 2020 and quickly found a large audience – thousands of researchers worldwide adopted it, and it has been cited more than 5000 times in the three years it has been operational. A new upgrade, TBtools-II, has now been developed, with more than 100 new features to make bioinformatics analysis easier than ever.
In recent years, bioinformatics analysis has become a mainstay of academic research worldwide – with new advances in biotechnology, it has become possible to extract a large amount of biological data from various sources. While this data is often instrumental in uncovering new insights, the quantity makes it impossible to analyze. Further, the variety of bioinformatics tools needed to clean and process the data as required can be numerous and daunting, not least because of different tasks requiring entirely different workflows. Valuable time is lost due to researchers being forced to learn to adapt to different platforms and interfaces before any analysis can be performed. Especially for researchers who work primarily in wet labs and may not have the coding proficiency required to operate these tools, such a lack of accessibility presents a significant challenge.
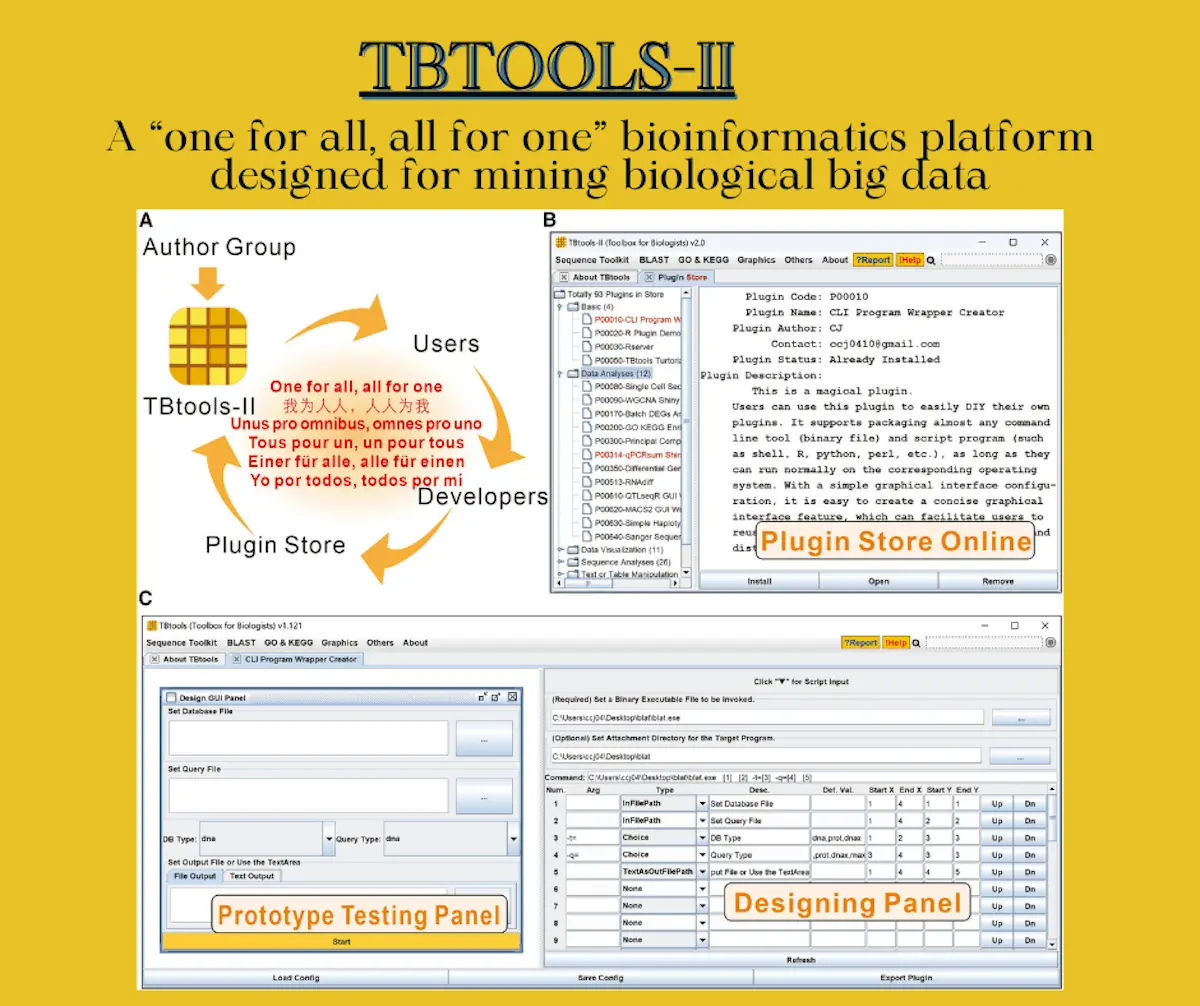
In 2020, the release of TBtools provided researchers with a viable solution to this problem: featuring more than 130 functions and receiving frequent updates and bug fixes, the toolkit has become ubiquitous in research labs. Despite its utility and functionality, the various needs of different users presented a significant challenge to the developers. Bioinformatics data analysis encompasses a wide variety of applications and tasks, and researchers working in certain fields require highly specific and personalized tools for data analysis. While the addition of these tools helped increase the usefulness of TBtools and helped it serve a wider section of researchers, it also bloated the toolkit significantly and made it harder to use and navigate.
This resulted in the need for a successor toolkit that incorporated the various developments made in the bioinformatics field over the past few years while remaining user-friendly and customizable. This also had to be achieved without bloating the application so much that it became difficult to distribute and install.
Feedback from users was taken into consideration when incorporating new tools and improvements for existing features: new tools were included for topics varying from comparative genomics to graphics configuration and phylogenetics. A new function, BLASTZone, has also been developed, allowing for concurrent querying with multiple BLAST databases. Users can manage and store databases in BLAST, and multiple databases can be selected simultaneously to BLAST target sequences against. A function for constructing phylogenetic trees has been incorporated into the tool, allowing for rapid visualization and interpretation of BLAST results for a given sequence. To further aid in phylogenetic studies, a new tool has been released – “Find the Best Homology” – which identifies homologous genes in a particular species for its target gene and can do so in a matter of seconds. This is a significant improvement over the basic BLAST tool, which only sorts homologs on the basis of e-values, as well as other companion BLAST tools for phylogenetic analysis, such as Orthofinder, which require more processing time.
To further streamline the task of constructing phylogenetic trees (which usually necessitates multiple steps such as sequence alignment, trimming, and tree reconstruction, among others), a tool called “One Step ML tree” has been created, which incorporates various separate tools such as MUSCLE, trimAI, and IQ-TREE2, and automates the workflow, allowing for easy visualization of the phylogenetic tree. Various advanced calculation algorithms for the evaluation of selection pressure in gene selection have also been implemented.
TBtools-II also improves on its predecessor’s capabilities regarding data visualization, allowing the use of pre-existing graph templates for the preparation of plots, as well as supporting the import and export of plot parameters files, allowing users to use the same parameters previously defined. The instant archive function also allows for the saving and sharing of intermediate files to different workstations, making it suitable for large-scale visualizations like Circos plots.
Despite the widespread applicability of these functions, the wide variety of analysis tasks present in the field means that it is impossible to accommodate the demands of every user due to size and distribution constraints. A new plugin mode has been developed in recognition of this that allows for various specific tools to be implemented in the form of dispensable plugins, which can be added or removed according to the needs of the users. Users are also encouraged to contribute tools, scripts, and pipelines so the community can use them. In order to assist users who aim to distribute their tools in this manner, a plugin has been created that allows users to transform nearly any kind of software or script into a plugin that is easy to access and use.
Conclusion
The motto of the TBtools developer team is “one for all, all for one” – a phrase that aptly describes the platform’s aim at distributing essential tools to bioinformaticians while utilizing the expertise of their userbase. The new plugin mode enables users to have an even greater degree of customizability, allowing for the personalization of the toolkit to match the demands of a particular project. Creating tools that allow for the easy packaging of pre-written software into a TBtools-compatible plugin also helps users share their tools with the community effortlessly. As local software, it is also more convenient to operate than web-based software, especially in regions that suffer from low or unstable connectivity. As software that serves as a versatile component of a researcher’s toolkit, TBtools-II provides a user-oriented and dynamic platform for bioinformatics tasks.
Article Source: Reference Paper | TBtools-II is freely available to non-commercial users at GitHub | Demo data is freely available at Website
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.
















[…] Demystifying Big Data with TBtools-II: A Versatile and Customisable All-in-One Platform for Bioinfor… […]