With the rise in popularity of the field of bioinformatics in recent years, researchers have created multiple computational tools that help streamline and optimize the research process. However, while the use of such tools may seem intuitive to an experienced researcher, newer researchers and researchers who don’t have as much experience with bioinformatics workflows may be turned off due to a lack of knowledge of how to use these. A new deep learning model, BTR (Bioinformatics Tool Recommendation System), has been developed to recommend the most suitable tools for a given workflow.
The creation of different workflows can be a challenging task as it requires a comprehensive grasp of the various computational tools that are available, as well as how to use them. Many workflow management programs have emerged to remedy this, with some of the more popular ones being Galaxy and Snakemake. They utilize a toolbox that consists of a set of functions and optimize the use of resources such that the entire process is efficient and easy to understand. However, new workflows may be necessary, especially in novel and less-explored areas, making these programs less useful. Many researchers may find it more useful to create an entirely new workflow that is modified to be optimal for their experimentation and research, but a lack of knowledge regarding the availability of tools for various functions serves as a deterrent.
It is obvious that this is an extremely time-consuming process, which may not be feasible for most researchers who have to work under strict deadlines and who may be prone to human error. Automated methods were thus developed especially for this purpose. EDAM and APE (Automated Pipeline Explorer) serve as indexes for various computational tools, which can be searched to find ideal tools. Other programs like WINGS and the Galaxy tool recommender system can also be used for this purpose. Even so, the process of actually constructing a workflow is still manual and can be extremely tedious to validate. It also lacks the specificity that is necessary for greater utility. The majority of these tools served one of two purposes: either the identification of certain implementations or the abstract definition of a workflow. A tool that successfully integrates both these aspects into a single program would be beneficial and would improve efficiency greatly.
In addition, many of these programs tend to be quite simplistic and reduce computational workflows to mere steps in a linear progression instead of the more branched and complicated versions that are more accurate depictions of the workflow.
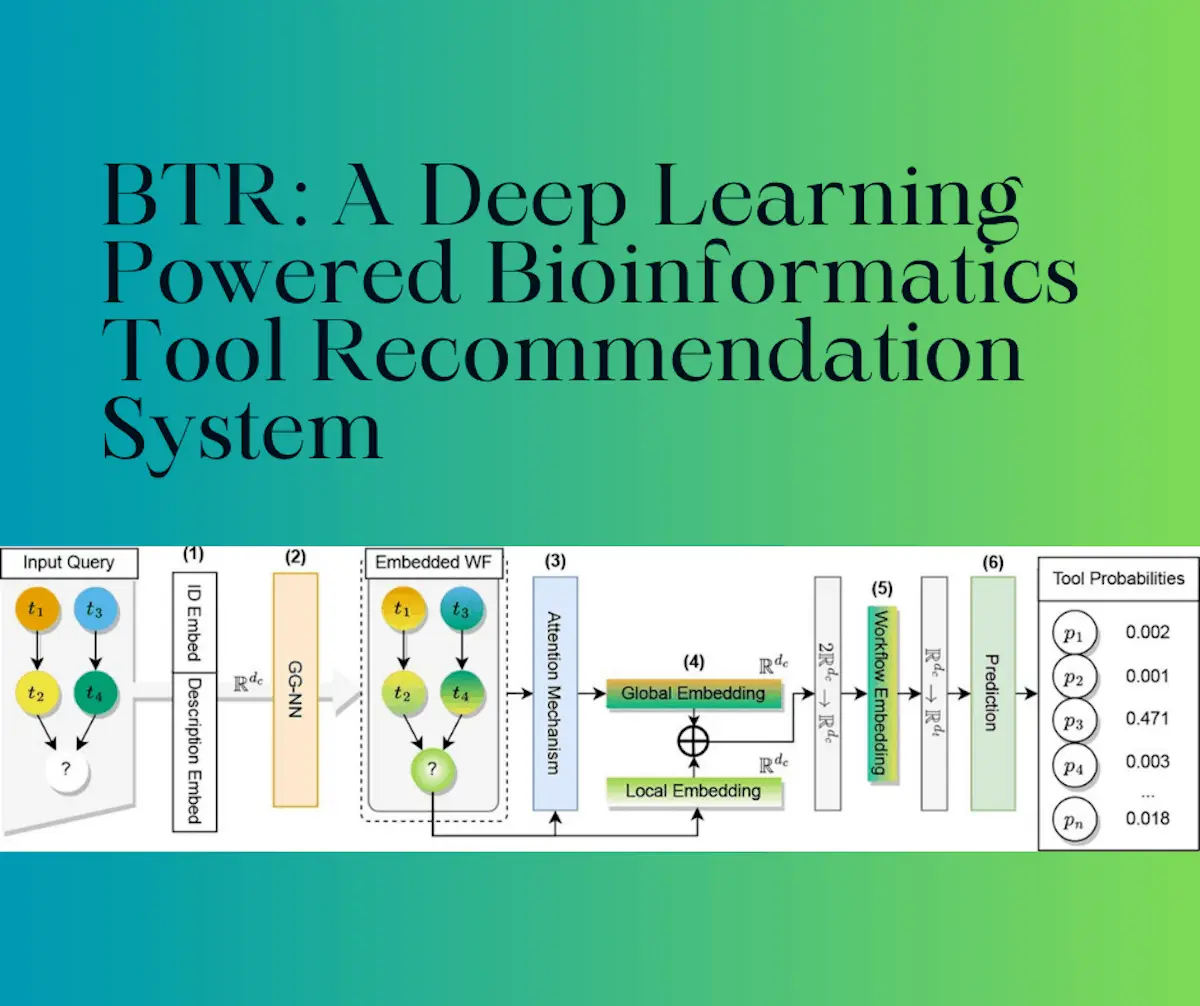
The Bioinformatics Tool Recommendation System was developed with the aim of fixing these issues. The workflow is constructed and modeled as session-based recommendation problems, and emergent graph neural network technologies are utilized in order to produce a graphical representation that accurately depicts the complex structural and functional context of the workflow. The workflow is thus represented as a directed graph.
The Bioinformatics Tool Recommendation System, as well as some variants, were evaluated in comparison with two different Galaxy databases, all of which combined contained more than 7000 workflows. The tool was then compared to a conventional baseline method. The BTR consists of a large toolbox comprised of many varying functions or ‘tools.’ The workflow was defined here as the “execution sequence alongside input and output connection between bioinformatics tools in order to perform a specific bioinformatics task.”
The task of tool recommendation was framed as a kind of regression problem in order to predict the tool that was most likely for a given workflow. Natural language processing methods were then applied in order to produce information regarding the various tools using their descriptions.
Once the final representation is produced, the likelihood of various tools is calculated, and the recommendation proceeds using this as the basis. The program was then evaluated by utilizing two datasets, which were collected from Galaxy workflows. Several variants were trained using different configurations and then compared their performance using various parameters with the aim of understanding the utility of the system for the purpose of constructing workflows specifically.
When assessing the results of the evaluations, it was found that the version of BTR that used graph representations for its workflows significantly outperformed the version that used linear sequences. It also outperforms the Galaxy Tool Recommender, which was used as a baseline, by over 50%. Case studies were then conducted to demonstrate the utility of the tool in various scenarios, including transcript assembly, single-cell analysis, and COVID-19 variation analysis. In all three cases, the model provided relevant recommendations despite limitations in the given data as well as an inability to understand user intent. BTR was then compared to the capabilities of the popular ChatGPT model, where it was found that the latter failed to provide answers that were as specific or as relevant as BTR by virtue of it not being optimized for that purpose.
Conclusion
It is plain to see that a tool that can construct workflows in this manner while requiring very little input is an attractive proposition for most researchers. This program also significantly outperforms other alternatives. The architecture can hence be used as a lone program or can be integrated into existing systems like Galaxy.
The system can also be modified as necessary to integrate various parameters and configurations to produce the most optimal output. One of the limitations of the software is the current paucity of datasets that can be used to train the model, but this should be rectified as more data is acquired. Additional options for user input could also significantly increase its applicability to different research requirements. Despite these limitations, however, the Bioinformatics Tool Recommendation System is a powerful deep-learning tool that can be added to the arsenal of bioinformatics researchers as they seek to optimize and improve their workflows.
Article source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.