Protein engineering is a fast-growing area of biotechnology. Responsible for carrying out nearly every biological function, the question of how to reliably engineer them in order to perform certain tasks intrigues scientists worldwide. So far, most attempts have focused primarily on reproducing the structures of wild-type proteins to use as a blueprint for novel protein design. Though this approach has been quite popular due to the relative ease of constructing new proteins, the reliance on already-existing proteins restricts the kind of structures these proteins can have. The study published in Nature Communications delves into the design of deeply knotted proteins, offering valuable insights into the folding mechanisms of knotted proteins
De novo design of proteins can result in the creation of protein sequences and folds that are unlike those that can be observed in nature. Until now, de novo design has been relatively simple. However, the challenge of constructing more complex structures can have a significant impact on the field by opening the door to more creative and complicated approaches.
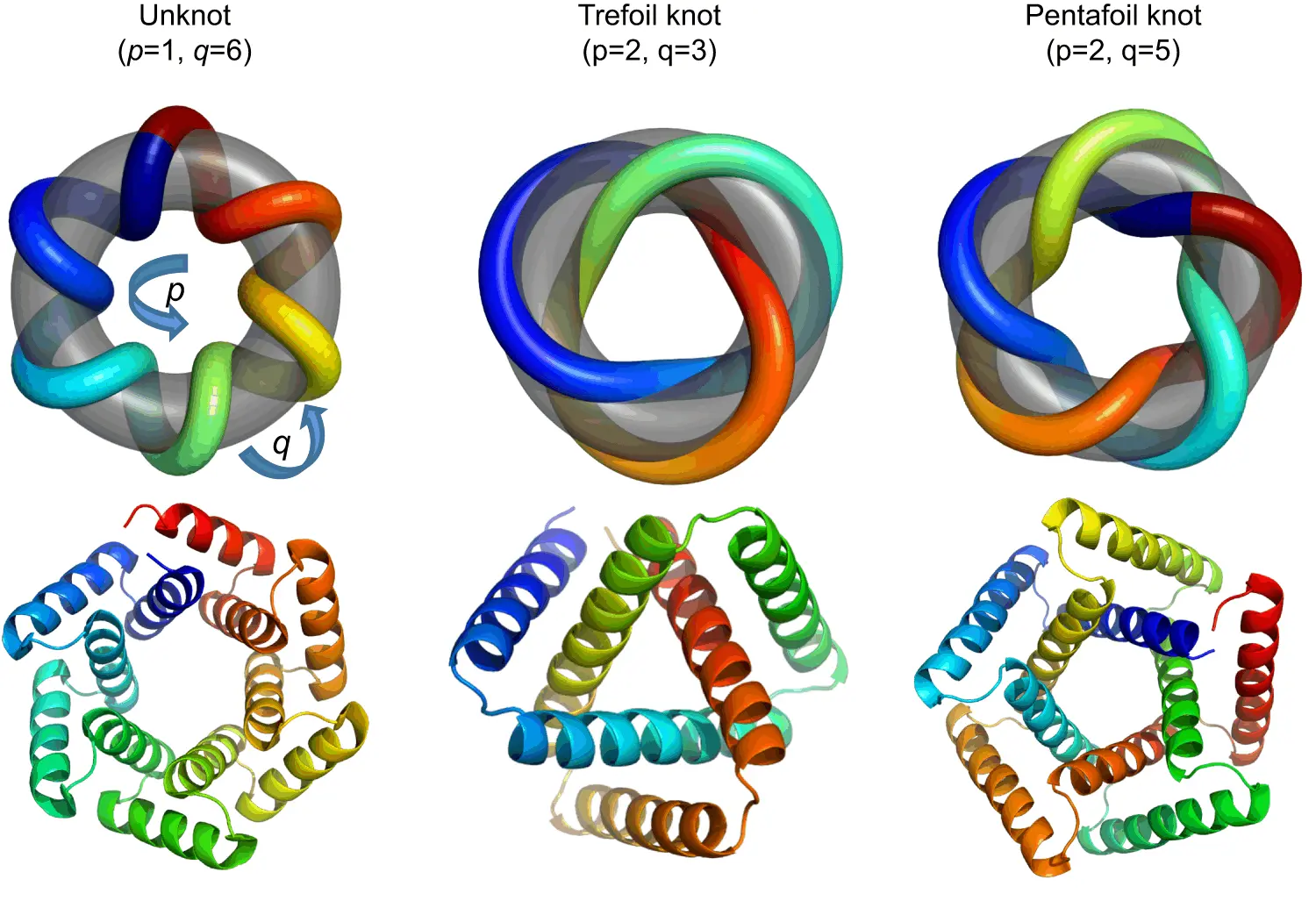
The design and characterization of knotted proteins is one such challenge: first conceptualized in the 1990s, they are still not properly understood, though more knotted proteins have emerged. These only form a tiny proportion of protein structures that are known to science, and their high diversity means that there are not many conserved features that can be used as a basis for the de novo design of these molecules.
Circular tandem repeat proteins are a diverse family of proteins of varying sizes and structural complexity. Tandem repeat proteins have extremely modular components, with an array of various structural motifs, which demonstrate themselves to be of particular interest for de novo designs. As of today, there have been no naturally occurring proteins found that are helical TRPs and show knotted topologies.
Four trefoil knotted proteins were designed initially, which corresponded to three different architectures. The termini of these proteins were located close to each other, resulting in a near-perfect symmetry. Of these four proteins, only three were expressed when transformed into E.coli cultures, and of these three, two proteins could be purified easily, though they couldn’t be crystallized. In an attempt to remedy this, additional modifications were made in order to change the molecule’s surface entropy in order to encourage the formation of crystals. These proteins were similar to their predecessors in behavior. A single protein from this generation was selected for further research. It was concluded through the use of atomic force microscopy that when unfolded, the remnant knot would be tightened in the unfolded peptide chain. This may have an impact on the elasticity of polymers and may even result in the backbone prematurely disintegrating through hydrolysis. However, such deviations were not observed in the course of the experiment.
It was also found that the protein’s tensile strength was much higher than other such proteins that comprised purely of alpha helices. It was noted that the unfolding force measures were much weaker than other such proteins like Titin IG27 or even Top7. The cause of this increased tensile strength is a matter of some contention: though computational experimentation has suggested that the knot contributes to this, testing in the lab revealed that knotted proteins did not demonstrate significantly higher tensile strengths than their unknotted counterparts. Another probable cause of the high tensile strength could be the presence of optimized non-covalent bonds.
The thermodynamic stability of the three proteins was then assessed at equilibrium, and these were found to be highly stable in their native states, with their half-life being around 100 years. It was also noted that the loss of alpha-helical structures in the intermediate construct, suggesting the presence of hydrophobic interactions, may help stabilize the intermediate. The protein was also shown to be stable at lower pH.
The Creation of Pentafoil Knotted Proteins
Bolstered by this success, a more complicated topology was taken as a new challenge: the pentafoil protein, which hasn’t been observed in nature at all. Three initial designs were constructed and characterized, though this proved to be much more difficult than it had been for the trefoil proteins due to a significant loss of sample when concentrated. Attempts at crystallization also proved to be unsuccessful. A second set of proteins was designed using ProteinMPNN, with their sequences modified, and these were filtered with the help of AlphaFold2 (which was helpful in generating models of the proteins despite them being designed de novo). Of these, five were cloned into bacterial vectors, and three were expressed successfully and at high concentrations.
Though it has been postulated for years that deeply knotted proteins did not exist due to the necessary interactions clashing with established rules of protein folding, it was discovered that multiple such structures did exist in nature and did so in an array of different structures. However, their complexity makes them intimidating to researchers in the field of protein design. However, the successful creation of knotted proteins de novo paves exciting new paths in the field.
The study validates the results of previous experimentation, which suggested that proteins were capable of folding to a knotted state without the need for evolutionary pressure. However, the increased difficulty of designing a pentafoil knotted protein in comparison to the trefoil knotted protein suggests that designing such proteins may be exponentially more difficult with greater complexity. Though the pentafoil proteins could not be successfully generated, as the only ones that could be expressed and characterized misfolded, resulting in a much lower contact order than was originally intended, further experimentation may result in its successful creation in the lab.
Conclusion
The designed proteins were compared to naturally occurring trefoil proteins that were found in the KnotProt database, allowing for an analysis of divergences in structure and properties. This resulted in the revelation of significant insights into the structural characteristics of these proteins and how they can vary between different protein families. Though further research is necessary to determine whether pentafoil knotted proteins can be successfully expressed and purified, the design of such proteins de novo has exciting implications for the field of protein engineering and biotechnology as a whole. Without the need to be constrained by the structures found in nature, new avenues for protein design can be explored.
Article source: Reference Paper
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.