New avenues are being paved in the world of healthcare every day. One of the most promising new developments is the creation of RNA-based medications. Demonstrating great therapeutic potential, this new class of therapies offers novel ways of curing diseases. However, their full potential hasn’t yet been unlocked, largely due to a lack of understanding of RNA structure and the various properties it possesses. Since the determination of structure through experimental means can be expensive and time-consuming, it proves disadvantageous in terms of practicality. The development of powerful new computational models, like ATOM-1, can potentially change this.
The development of RNA-based medications and therapies has largely been hampered due to the lack of information available regarding its structure, functions, and properties. Due to a lack of data, it is also difficult to model computationally, and very few publicly available RNA structures are seen in databases such as the Protein Data Bank.
Machine learning has been successfully utilized to create accurate results using limited data. This is facilitated by the use of transfer learning – models trained on large sets of data allow for better and more precise predictions. Probe networks are a technique commonly used to depict the creation of various internal representations. These small neural networks utilize internal embeddings taken from the pre-trained model as input to produce accurate predictions of molecular properties. Theoretically, a foundation model that possesses a large set of internal representations of various RNA structures would facilitate the creation of accurate predictions even for tasks where the amount of data available is severely limited. It’s important to note that training such foundation models necessitates using a dataset that is appropriately large, diverse, and informative.
Chemical mapping is an efficient method for gaining a large RNA dataset: through the use of chemical reagents, RNA nucleotides can be modified as desired in a manner that is dependent on the molecule’s overall structure, and different reagents and conditions will result in various aspects of the structure being measured. The modifications can then be identified through RNA sequencing, which, combined with next-generation sequencing, allows for a quick and accurate method of gaining large amounts of data. In addition, this method provides an advantage compared to the baseline method (which involves multiple sequence alignment) as it can be used on arbitrary sequences of RNA, facilitating research on sequence space. Since such experiments provide a direct measure of structural information, foundation models trained on this data may provide more accurate predictions than conventional computational models.
The probe network is then constructed using this data. The data is collected in a set of libraries, and a custom computational model is fit to it. A probe is first trained such that it is able to provide predictions on various properties using the embeddings from the foundational model. Further, to demonstrate that the probe’s predictions are due to the foundational model instead of other factors, another version of the network is trained with restricted access to the internal embeddings. Through a comparative study, it is possible to determine whether the former performs better than the latter and, if so, whether it is due to the influence of the foundation model that the predictions made are more accurate.
ATOM-1: A New Method for the Prediction of RNA Structure
ATOM-1 is a new foundational model created to address this need: trained on data obtained through in-house chemical mapping experiments, this model possesses a rich array of internal representations. While small, the probe networks in the model demonstrate great accuracy in tasks such as the prediction of 3D structures secondary structures, as well as the stability of RNA, outstripping other methods by a large margin.
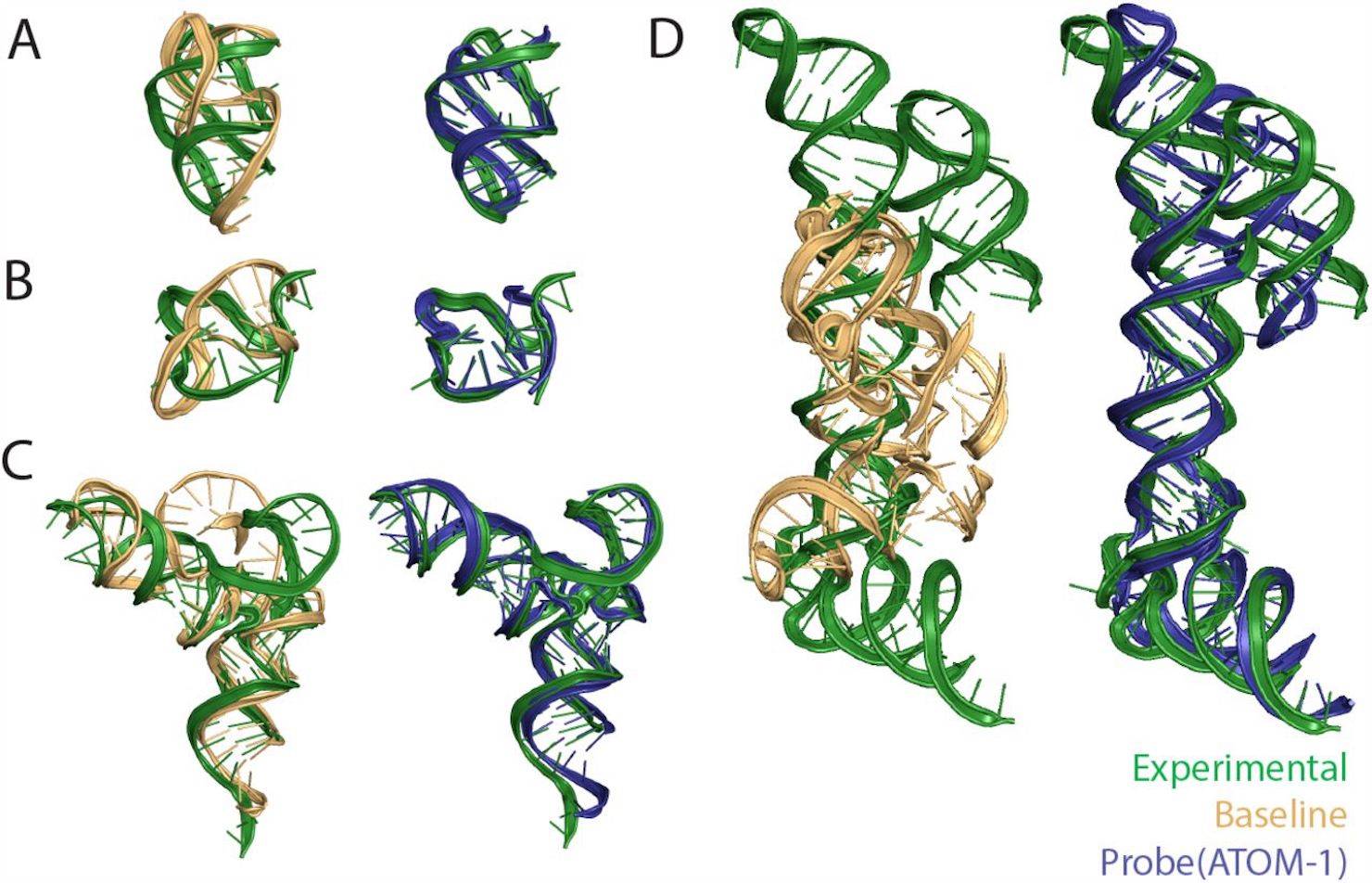
The probe trained on ATOM-1 was found to generate predictions of great accuracy for various kinds of complex RNA structures from diverse classes. The probe was also consistently capable of predicting the presence of various elements of the secondary structure, such as pseudoknots, which more conventional methods are unable to do. The proposed model, trained on structures obtained from the Protein Data Bank, was used to predict tertiary structure elements. The results of its predictions were then compared to those obtained from different structure prediction software. It was found that despite its lack of access to data from MSAs and its being smaller and shallower, the probe did have greater accuracy than RoseTTAFold2NA and RhoFold (when measured using the RMSD method). It also had consistently better accuracy when compared to the baseline network, which didn’t have access to the embeddings developed in ATOM-1.
The ability of the model to predict RNA stability was also measured, with the probe’s predictions of reactivity and degradation characteristics being more accurate than competing models when participating in a community challenge centered on the task of stability predictions.
Conclusion
It is seen that ATOM-1 possesses a strong capacity for generalization, suggesting its suitability for various applications in the realm of RNA therapy development, such as siRNA toxicity, ASO activity, and translation efficiency. When provided with a small dataset, the probe is able to provide fast and accurate predictions, allowing for greater efficiency in research while also lowering costs.
Currently, only probe networks have been utilized with ATOM-1, but more extensive networks that use advanced techniques for transfer learning may have significantly better accuracy. While trained solely on chemical mapping data, the addition of other relevant data to the datasets used to train the foundational model can have a positive impact on the network’s accuracy and generalization abilities.
Article Source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.