Tumors that affect the central nervous system are among the most fatal kinds of cancer, especially in pediatric cases. Surgical removal of the tumor is usually the prescribed course of action, made harder by the fact that the surgeons performing the procedure will not be able to determine the kind of tumor they’re dealing with beforehand, limiting their ability to come up with a plan in advance. A new tool utilizing neural networks seeks to improve the landscape of cancer diagnostics by providing a rapid and accurate mechanism to classify CNS tumors in an intraoperative setting.
Primary treatments for tumors of the nervous system usually involve the excision of the tumor through surgery, this is a difficult procedure to perform, requiring surgeons to make decisions regarding how much of it can be excised without posing a threat to the neural system and increasing the chances of comorbidity.
The determination of the kind of tumor is crucial to be able to identify the necessity and amount of resection that can be feasibly carried out. Certain tumors are presently viewed as being incurable, hence, the surgeon’s aim while operating on a patient must be to acquire tissue samples for proper diagnosis while preserving the patient’s quality of life as much as possible. However, it may be beneficial to attempt a complete resection as possible for other kinds of tumors, as it improves survival.
The lack of prior knowledge regarding tumors means that patients often have to undergo surgery again if they are unable to have a complete resection when one is needed, or it may be found later that a more cautious approach would have worked equally as well. Both of these outcomes may hinder the patient’s quality of life and necessitate further invasive procedures, increasing the risk of resulting infections and stress in the body. Hence, for accuracy and taking into account an individual patient’s needs, it is beneficial to have a reliable way of diagnosing tumors.
Cancer diagnostics methods can take various forms: one way is by assessing methylation patterns within the tumor. Often, certain kinds of tumors will have distinctive and similar patterns of CpG methylation, allowing for accurate conclusions about the tumor’s origins and prognosis. This is often done through the use of machine learning tools, and methylation arrays are commonly utilized for this purpose. However, this time-consuming process requires several days to complete, making it unfeasible for intraoperative approaches, where only one surgery has to be performed.
A new method called nanopore DNA sequencing allows for sequencing-based diagnosis at a rapid pace. Its low costs, form factor, and instantaneous availability of data are some of its major advantages compared to older methods. This method can directly measure methylated cytosines, significantly reducing the time needed to prepare samples. This process is so fast that it is possible for tissue samples to be acquired sequenced and for a diagnosis to be obtained within a short amount of time, such that the surgery can proceed with newly acquired information regarding the tumor type. However, it is not known beforehand what CpG sites will be used, and the short amount of time available means that the generated methylation profiles will be quite sparse.

The Development of A New Tool for Cancer Diagnostics
A new approach has been developed that enables the classification of tumors within intraoperative settings – Sturgeon is a patient-agnostic neural network classifier optimized to work with very little data. Using this approach, neural networks were painstakingly trained on more than 36 million simulated nanopore runs and were then validated on 4.2 million more data samples. This extensive validation and testing allow Sturgeon to classify samples based on very sparse data accurately – this is to simulate the limited capability of nanopore sequencing methods within the short timeframe between sample acquisition and diagnosis, allowing it to work in an intraoperative setting.
The Sturgeon models were then applied to 50 samples from CNS tumors as well as 514 nanopore-sequenced samples. Out of these, 45 samples out of 50 were correctly classified, even with the sparse data available. Additionally, there is a paucity of the kind of datasets required for training these neural networks – to remedy this, Sturgeon utilizes data augmentation to simulate a large number of unique sequencing data samples from every methylation profile it uses. Thus, 94 profiles from a varied group of patients were obtained, and 500 sequencing experiments were simulated at seven depths to give a total of more than 300,000 samples. The Sturgeon classifier was then applied to these samples, where more than 95% of cases with clear diagnoses were accurately classified, showcasing that an accurate diagnosis can be achieved with sparse data with low rates of error. The vast majority of the misclassifications that occurred were from simulated experiments derived from only two samples, both from the same family. When tested on data obtained from pediatric tumor samples, accurate classification was achieved (with a confidence score of more than 95%) around 77% of the time, and high-confidence misdiagnoses were reported only 0.03% of the time. In a publicly available dataset, it was able to classify 92% of the available samples correctly.
A downsampling method was also used to detect large copy number variations, though smaller ones weren’t detected as reliably. Sturgeon was then used during the course of 25 surgeries being performed in the Netherlands and was able to classify 72% of tumors correctly using only 45 minutes worth of sequencing data.
Sturgeon’s accuracy was significantly lower in those cases that had been deemed difficult to diagnose, with only 11 of 26 total samples being accurately classified. This is a common limitation of machine learning models: accurate conclusions can be reached only if the samples have been represented in the data that they were trained on. In addition, it is not yet known whether the accuracy is affected by other factors like sample purity. Sturgeon can also be used in tandem with adaptive sampling, though this requires specialized hardware.
Conclusion
Sturgeon’s quick and precise diagnoses make it possible to provide better care to patients with CNS tumors and to analyze and modify the surgical strategy within an interoperative setting. It can be advantageous in cases where histological conclusions are ambiguous and can help medical professionals make better decisions with regard to their plan of treatment. Sturgeon proves to be an exemplar of the power of cross-specialization innovation, as the power of neural networks and biological knowledge combine to provide efficient and accurate cancer diagnostics tools with the potential to improve the lives of patients around the globe.
Article source: Reference Paper | Reference Article
Learn More:
Sonal Keni is a consulting scientific writing intern at CBIRT. She is pursuing a BTech in Biotechnology from the Manipal Institute of Technology. Her academic journey has been driven by a profound fascination for the intricate world of biology, and she is particularly drawn to computational biology and oncology. She also enjoys reading and painting in her free time.











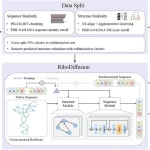




[…] in the Netherlands created an A.I. system called Sturgeon. It uses a patient’s tumor DNA to diagnose the tumor type and subtype. This gives the surgeon […]