The University of Alabama in Birmingham researchers have found 16 unique cell populations in a complex part of the midbrain known as the ventral tegmental area, or VTA, using single-cell RNA sequencing in a study that advances the field.
Expanding our understanding of the VTA’s function will help us better understand the mechanisms behind substance use disorders like cocaine, alcohol, opioids, nicotine, and psychiatric illnesses like schizophrenia and attention deficit hyperactivity disorder, or ADHD.
The VTA is significant because of its role in reward-directed behavior’s dopamine neurotransmission. Dysregulation of these reward pathways occurs in substance use disorders, resulting in drug craving despite adverse consequences. In the most recent year, more than 100,000 drug overdose deaths were reported in the United States. The VTA is also involved in several other neuropsychiatric conditions.
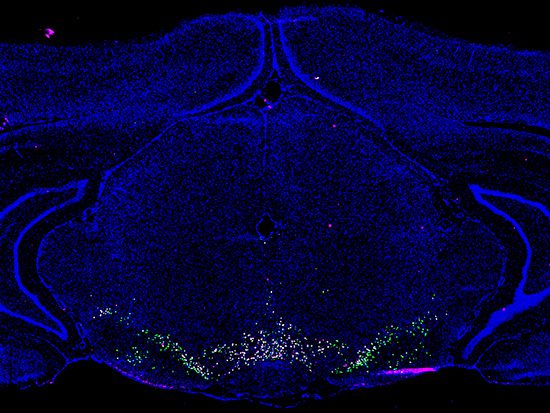
Image Source: https://www.uab.edu/news/research/item/12778-r-i-a-key-brain-region-for-substance-use-disorders-now-has-a-searchable-atlas-of-distinct-cell-populations
Dopamine is a neurotransmitter that helps the brain relay impulses between nerve cells by acting as a chemical messenger. While decades of research have concentrated on dopaminergic neurotransmission in the VTA, there is now strong evidence that GABA and glutamate, two additional neurotransmitters functioning in the VTA, are important in reward-related behaviors. There’s also evidence of “combinatorial” neurons, which may produce and release various neurotransmitters. These findings point to a new level of complexity in the cellular and synaptic function of the VTA.
Systematic biology is the science of categorization, and it usually relates to the classification of species based on their natural relationships. The UAB VTA project classifies cell populations to extend and refine prior work on the many cell types in the VTA and provides a starting point for comprehending the interactions between these cells and their vast connections to other brain areas.
Single-nucleus RNA sequencing of 21,600 cells from the rat VTA led to the identification of 16 unique cell types by changes in gene expression, resulting in a searchable online atlas of the VTA. The rat is the most commonly used model in reward and substance abuse research. This unbiased approach was utilized to construct the largest and most thorough single-cell transcriptome investigation focused only on the composition and molecular architecture of the VTA, in contrast to earlier research that picked particular subgroups of cells for RNA sequencing.
Even though the VTA is made up of various cell types, the majority of studies on the subject have relied on more focused techniques like immunohistochemistry, in situ hybridization, and single-cell qRT-PCR. Though it was already known that the VTA is made up of various cell types, the UAB atlas adds to existing studies in a number of ways.
“For example, previous single-cell sequencing studies were conducted exclusively in the mouse brain and have relied primarily on sequencing a subset of fluorescence-activated cell sorting-isolated midbrain dopaminergic populations, rather than sampling all VTA cell types,” says the corresponding author of the study Jeremy J. Day, Ph.D. “Notably, our sequencing dataset focuses exclusively on VTA sub-regions, unlike other studies that have focused on pooled cells from the mouse substantia nigra and VTA or a subset of fluorescently tagged cells from general midbrain regions.”
The researchers used an unbiased method to conduct transcriptome profiling of all cell types in the VTA, resulting in the identification of 16 transcriptionally different cell populations and several neuronal subtypes. Classic dopaminergic neurons, three glutamatergic neuron subsets, three GABAergic neuron subsets, and nine other cell types, including astrocytes and glial cells, are among the 16 different cell groups.
Image Source: An atlas of transcriptionally defined cell populations in the rat ventral tegmental area
Using single-cell sequencing, DEG testing can discover markers for viral targeting of different VTA DA (dopamine) neuron subtypes.
Only 399 nuclei out of 21,600 possessed gene markers for dopamine (DA) neurons that had previously been identified. This number of DA neurons reflects a previous scRNA-seq study that revealed 991 DA neurons in a combined substantia nigra and VTA sample of 44,091 cells.
A gene encoding the enzyme guanosine triphosphate (GTP) cyclohydrolase 1 (Gch1) was found as a selective marker of classically characterized DA neurons using differential expression analysis. Gch1 is a rate-limiting enzyme in tetrahydrobiopterin synthesis that also serves as a cofactor for TH and is required for the generation of DA. As a result, cells missing Gch1 may lack the machinery required for de novo DA production.
The UAB researchers detected four sub-clusters after sub-clustering neuronal cells, which could represent neurons capable of combinatorial neurotransmitter release. They also discovered selected gene markers for both traditionally defined and combinatorial dopamine neurons. A selective marker permits viral targeting of different VTA subtypes for functional research.
The researchers then looked at sub-clusters for opioid neuropeptides and receptors, finding pan-neuronal enhanced expression of risk genes linked to schizophrenia and “smoking initiation,” as well as enrichment of attention deficit hyperactivity disorder (ADHD) risk genes in two glutamatergic neuronal populations.
Story Source: Phillips III, R. A., Tuscher, J. J., Black, S. L., Andraka, E., Fitzgerald, N. D., Ianov, L., & Day, J. J. (2022). An atlas of transcriptionally defined cell populations in the rat ventral tegmental area. Cell Reports, 39(1), 110616. https://www.uab.edu/news/research/item/12778-r-i-a-key-brain-region-for-substance-use-disorders-now-has-a-searchable-atlas-of-distinct-cell-populations
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.











