To better comprehend the function of cells, scientists examine how different components of a cell – from single molecules to several organelles – work in synchronization. They studied individual molecules using classic structural biology tools, honing individual atoms. However, this method yields only static photographs of molecules in most circumstances. Instead, the scientists used cryo-electron tomography to determine how molecular structures react in their cellular environment. In contrast to other structural methods, this technique does not allow viewing atomic details.
Video Source: https://www.embl.org/news/science/observing-the-secret-life-of-molecules-inside-the-cell/
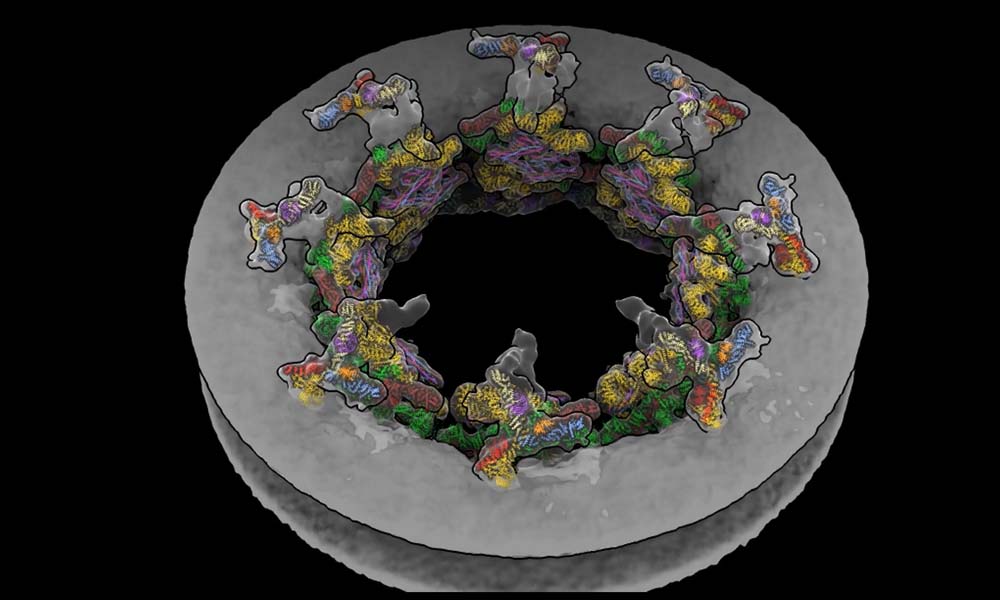
Thus, until now, it appeared impossible to observe the secret lives of molecules at the atomic level in their natural environment. This technological gap has been closed by a group of researchers from EMBL Hamburg’s Kosinski Group, the Beck Laboratory at the Max Planck Institute of Biophysics, and the Mahamid group, along with the Advanced Light Microscopy Facility at EMBL Heidelberg. They visualized how a huge molecular machine termed the nuclear pore complex (NPC) interacts inside cells using molecular modeling and cryo-electron tomography.
The NPC is a doughnut-shaped structure that contains a thousand proteins and is found in the nucleus membrane. It acts as a checkpoint, regulating which proteins enter and exit the nucleus and plays a crucial role in various cellular activities. Many viruses, including HIV and influenza, exploit the NPC to gain access to the nucleus.
The scientists were able to generate a visual that depicts how the NPC dilates and contracts in real cells in response to changes in the cellular environment by combining mathematical models with tomography data. Insights gained into this molecular behavior might aid in developing novel antivirals in the future.
Video Source: https://www.embl.org/news/science/observing-the-secret-life-of-molecules-inside-the-cell/
Without Assembline, a program created specifically for this task by the Kosinski Group, observing structural changes in such a huge complex at the atomic level would have been impossible. Assembline combines data from multiple experimental methodologies to allow structural modeling. It combines the features of numerous current tools into one tool, making it a ‘Swiss army knife’ for molecular complexes research. The program has previously proven effective in investigating other large complexes, such as the tuberculosis-related ESX-5 complex. It’s also free to use for the scientific community because it’s open source.
This research exemplifies EMBL’s approach to investigating life in context at several scales, which is at the heart of the future EMBL Program Molecules to Ecosystems. The EMBL is likewise committed to open research, and Assembline adds to the EMBL’s long tradition of making scientific materials freely available to the scientific community.
Story Source: https://www.embl.org/news/science/observing-the-secret-life-of-molecules-inside-the-cell/
Zimmerli, C. E., Allegretti, M., Rantos, V., Goetz, S. K., Obarska-Kosinska, A., Zagoriy, I., … & Beck, M. (2021). Nuclear pores dilate and constrict in cellulo. Science, 374(6573), eabd9776.
Rantos, V., Karius, K., & Kosinski, J. (2021). Integrative structural modelling of macromolecular complexes using Assembline. bioRxiv.
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar
- Dr. Tamanna Anwar