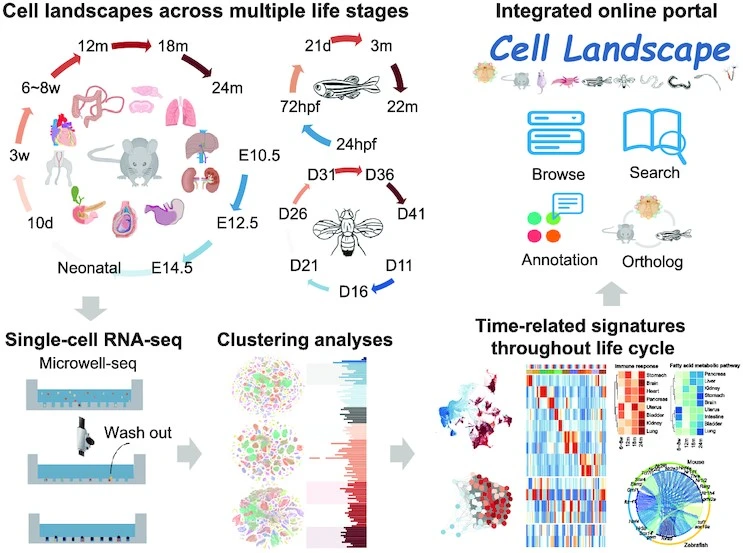
Life is based on individual cells. Landscape dynamics across species have not been compared despite extensive efforts to characterize cellular heterogeneity. A large number of cell landscapes were mapped for mice, zebrafish, and Drosophila at multiple life stages using single-cell RNA sequencing (scRNA-seq). With over 2.6 million single cells integrated, the construction of a cross-species cell landscape identified signatures and common pathways that changed over life. Activating mitochondrial metabolism pharmacologically alleviated aging phenotypes in mice by reducing structural inflammation and mitochondrial dysfunction. An integrated online portal – Cell Landscape – contains the cross-species cell landscape along with other published datasets.
In vertebrate systems, such as Xenopus, zebrafish embryos, human cell landscapes (HCL), Tabula Sapiens, mouse cell atlases (MCAs), mouse cell differentiation atlases (MCDAs), Tabula Muris, Tabula Muris Senis, cell atlases covering fetal and adult periods have been developed. There are also several invertebrate cell atlases available, including those for Caenorhaditis elegans, Drosophila embryos, Nematostella, sea squirts, fruitflies, and earthworms. As a result of these studies, science now has a far better understanding of how the cellular hierarchy functions in different species. RNA sequencing of single cells (scRNA-seq) offers unprecedented opportunities for reshaping knowledge about different types of cells. In the era of scRNA-seq, single-cell transcriptomic atlases are now possible at the organism level due to the dramatic increase in throughput.
Dynamic cell atlases, along with static atlases, will add another dimension to the understanding of life. Researchers have examined regulatory programs that govern the trajectories of cell fate decisions at the single-cell level during embryogenesis. The mechanisms of aging have also been studied using rodents. In these studies, immune cells were found to be significantly infiltrated and activated, transcription was heterogeneously expressed, and aging cells were found to have an unrestricted identity. In most cases, these analyses are limited to a few specialized systems and therefore are not comprehensive enough to compare different species and tissues. As a result of the universality of scRNA-seq, such comprehensive analysis can uncover both cellular and organismal-level rules that drive life progression.
A total of over 2.6 million single cells were profiled in this study from mice, zebrafish, and Drosophila at various stages of their lives. Through a cross-species analysis of these datasets, common pathways that change across life spans were examined. In addition to discovering the dynamics of gene expression and time-dependent trends in transcription factor (TF) regulation, the study discovered that cell types and genes tend to change with development and aging, particularly immune cells. The most common hallmarks of organism aging are structural inflammation and mitochondrial dysfunction, and structural cells exhibit evident inflammatory responses as they age. Activating mitochondrial metabolism pharmacologically also alleviated mice’s aging phenotypes.
Evolutionarily, developmental expression changes are conserved across multicellular organisms, and aging and development are highly intertwined processes. Among mice, zebrafish, and Drosophila, immune cell dynamics gradually changed with age in most tissues and cell lineages. All three species showed similar gene expression patterns throughout the life cycle: ATP metabolism and oxidative phosphorylation continued to decline, and immune responses were consistently activated. As a result of TF regulation analysis, it was found that some lineages of cells are regulated similarly across species by similar gene families.

Image source:https://doi.org/10.1093/nar/gkac633
During aging, structural cells in vertebrates demonstrate increasing characteristics of immune activation as the interaction between structural cells and immune cells gradually increases. Aged structural cells showed significant upregulation of immune-related genes, particularly those related to antigen presentation. Through the analysis of published datasets, both gene expression and epigenetic studies were further validated. Oxidative damage reduces mitochondrial function with age, according to the mitochondrial theory of aging. Invertebrates and vertebrates both exhibit rapid declines in genes involved in oxidative phosphorylation as they age. Inflammatory responses may also be closely associated with mitochondrial damage during aging. Cellular apoptosis, autophagy, and inflammation are known to be caused by mitochondrial dysfunction. Two-year-old aging mice were treated with PGZ, a drug shown to improve mitochondrial function.
As a result of treatment with PGZ, mice were able to perform better in exercise, had higher lipid levels, and had improved insulin response. PGZ treatment was shown to repair mitochondrial function and reduce structural inflammation in aged mice using scRNA-seq. The dose of PGZ used for the i.p. injection was relatively high (15 mg/kg/day) to achieve a pharmacological effect after 3 weeks of administration. Future clinical trials should consider long-term and low-dose treatments to reduce PGZ’s potential side effects.
At present, there are several types of single-cell RNA databases, but most of them are developed for a single study, a single tissue, or a single organ, and they only provide basic visualization and searching. Some are even difficult to access. While some integrated databases already exist, they tend to provide downloadable functionality and cell cluster visualization rather than interactive, precise search functions and prediction capabilities. Cell Landscape is an atlas-level database that provides not only a single-cell resource for representative animals’ atlases but also rich functions and useful tools for research on single cells. Cell atlases can be viewed in their entirety through the visualization function, and single clusters and marker genes can be accessed quickly. Annotating unknown clusters and finding potential markers can be made easier by searching for orthologs of genes of interest. It is easy to classify and annotate cell clusters in different species using single-cell mapping pipelines and cross-species mapping pipelines.
In conclusion, the study offers valuable insights and resources for studying organism-level development, maturation, and aging. Cell Landscape is a website that lets users browse single-cell landscapes of different species to facilitate the use of the data by the broader research community.
Article Source: Reference Paper | Cell Landscape Website
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.