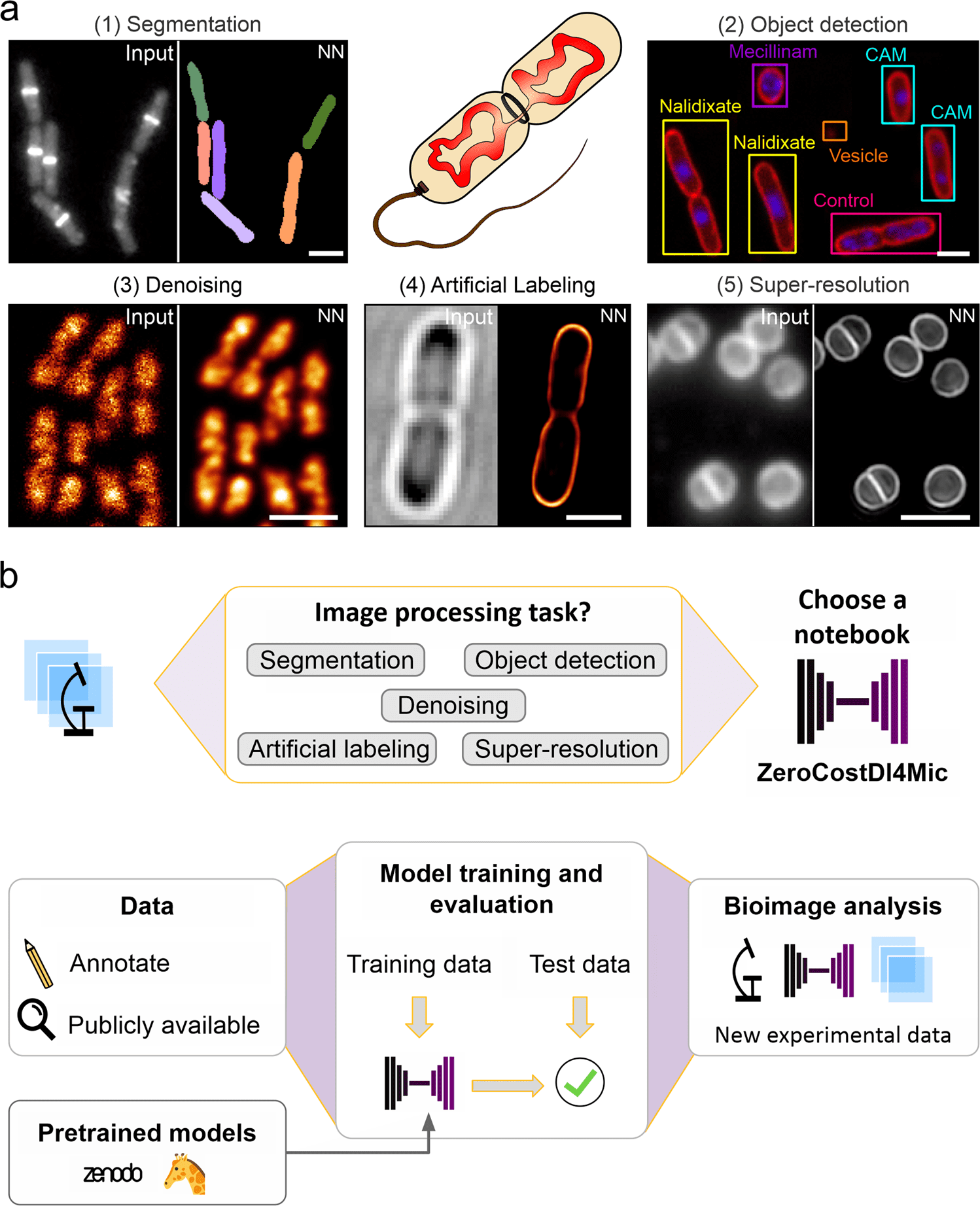
Scientists from University College London demonstrate the use of various cutting-edge artificial neural networks to analyze images of bacterial microscopy. The approach will help future researchers to explore the prospective deep learning (DL) applications and employ pre-trained models in their research on a designated platform.
Microbiology, the study of microorganisms and their community, is a multidisciplinary approach that combines various other disciplines, like biophysics, molecular biology, and biochemistry. It covers a wide range of spatial scales, from molecules to individual cells to entire ecosystems.
Image Source: https://doi.org/10.1038/s42003-022-03634-z
Increasing data volumes in microbial studies make classical data interpretation and analysis challenging, requiring more sophisticated computational approaches to retrieve relevant features from the data landscape. The use of machine learning (ML) is, therefore, increasingly replacing manual analysis of data.
As DL is highly versatile and fast, it has recently gained a lot of attention for bioimage analysis due to its high performance and speed. Image segmentation is closely related to object detection, which creates a bounding box and a class label for each detected object instead of classifying pixels as background or foreground. Self-driving cars or detecting objects in photographs are examples of real-life applications of this technology.
In addition to improving image quality, denoising images, and predicting super-resolution images from diffraction-limited images, this technology was also demonstrated impressively for image segmentation, object detection, and object classification.
Image-to-image translation and artificial labeling are two transformative applications created by DL tools. By simplifying their use and providing pre-trained models, novel DL approaches are being developed, and efforts have been made to make them available to non-experts.
A comprehensive note on DL models is that their training is highly dependent on the type of data used. The difficulty of generating models with good generalizing capabilities means that even slight variations in image acquisition parameters can render a model unreliable and unfit for further analysis.
Many parameters determine the quality of images, including different magnification (pixel sizes), variations of the focal plane, illumination patterns, a camera setting, and so on. Even if they fail to provide satisfying results, pre-trained models can be used for transfer learning, which speeds up training and improves performance.
Microbiologists heavily rely on DL approaches for segmentation, which enables automated analysis of large datasets and single-cell analysis in image analysis pipelines. It is possible to use such pipelines to automate counting cells or to analyze their morphological characteristics. Despite this, there is no universal DL network that excels at all types of data due to a wide variety of microscopy techniques and bacterial shapes.
The application of deep learning to bacterial images remains largely underexploited, although other applications such as object detection, denoising, artificial labeling, or resolution enhancement are well suited to this technique.
Smart imaging approaches, like automated image acquisition, can use object detection to determine the types and states of cells in microscopy. Live-cell imaging involves high contrast and fast image acquisition, which are necessary for fully capturing biology’s dynamic nature, making deionizing and artificial labeling particularly useful.
At times deionizing and artificial labeling often come with high illumination regimes, which often do not allow live-cell imaging to take place due to their high illumination power. Using robust denoising techniques for low signal-to-noise ratio (SNR) images reduces phototoxicity, increases temporal resolution, and allows shorter integration times, which reduces phototoxicity.
By using artificial labeling networks, it is possible to further reduce phototoxicity. Pseudo fluorescent images are created from bright-field images, histology images, or electron microscope (EM) images. The use of artificial labeling in live-cell applications is particularly useful for transforming bright fields into fluorescent signals. It provides molecular specificity while being even less phototoxic than denoising of low-SNR images since fluorophore excitation is not necessary.
The neural network creates virtual fluorescence images of structures or molecules imprinted by bright field images (e.g., membranes or nucleic acids) based on features in bright field images. The time required for data curation is reduced due to the absence of manual annotations in artificial labeling, which is contrary to image segmentation.
By training networks using bright field images as inputs, we can train networks for a wide range of subcellular structures, resulting in a high degree of multiplication. Based on this approach, multiple subcellular structures and their dynamics can be predicted in mammalian tissue culture samples.
Enhancing resolution is another way to increase the information content of microscopy images. Low-resolution images can be converted to high-resolution images using several supervised DL approaches. This includes confocal-to-STED, widefield-to-SRRF, and widefield-to-SIM transformations.
These networks also increase spatial resolution while reducing the amount of light required. As a result, live mammalian cells could be imaged in multicolor as well as cytoskeletal proteins, and endocytosis machinery could be viewed in high resolution computationally. However, the technology has not yet been applied to microbiology.
DL approaches already used to analyze eukaryotic specimens can easily be used to analyze bacterial bioimages to diversify the uses of DL technology in microbiology.
For successful DL training, a dataset with a wide range of images containing different bacterial species like Escherichia coli, Staphylococcus aureus, and Bacillus subtilis along with their imaging modalities including-bright field, widefield, confocal fluorescence, and super-resolution microscopy were generated.
Thus, this approach provides a designated platform that can be used by researchers and even non-researchers aspiring to be future researchers to explore potential Deep Learning applications and apply pre-trained models to their research.
Article Source: Spahn, C., Gómez-de-Mariscal, E., Laine, R.F. et al. DeepBacs for multi-task bacterial image analysis using open-source deep learning approaches. Commun Biol 5, 688 (2022). https://doi.org/10.1038/s42003-022-03634-z
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.