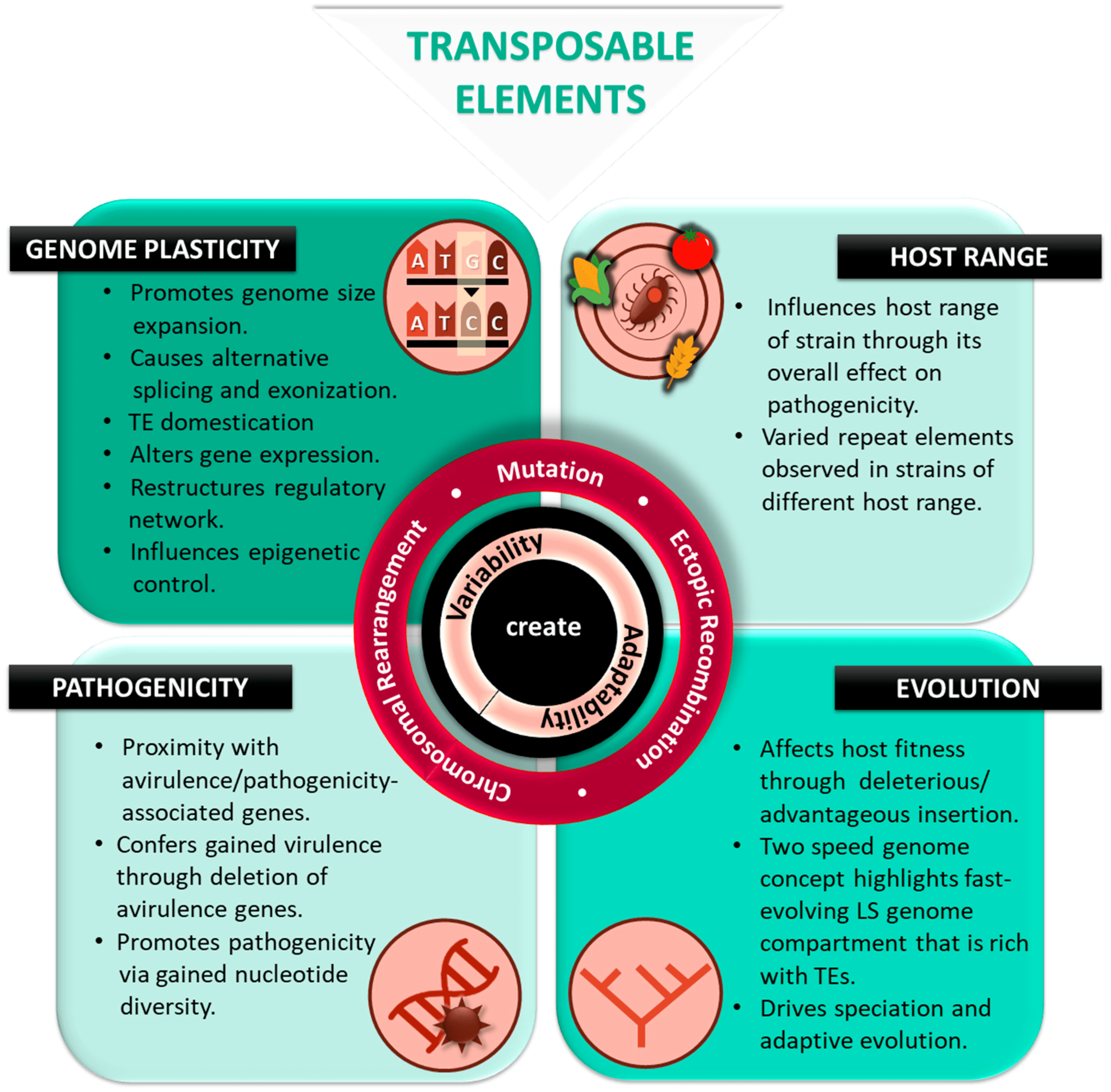
Scientists at the University of California, Irvine, provide new insights into “parasitic genes” or transposable elements. Despite accounting for more than half of human DNA, there is still much to learn about these elements. The new perspective could one day aid in the fight against cancer and aging-related diseases.
The transposable element (TE) is an aggressive genetic parasite that increases its copy number at the expense of the host’s fitness. There is a wide difference between species in the amount of TEs that are successful or how abundant they are across the genome. Despite the wide variation in TE abundance, evolutionary genomics has struggled to explain its origins. The spreading of repressive epigenetic marks left by silenced TEs into adjacent sequences might explain species-specific TE abundances.
A step-by-step analysis of this TE-mediated local enrichment of repressive marks, or ‘the epigenetic effect of TEs’, in six species from the Drosophila melanogaster subgroup was conducted to determine the importance of such effects in the abundance of genomic TEs. A significant variation across and even within species was found in the prevalence of local enrichment of repressive marks mediated by TEs. TE-mediated effects alter the epigenetic states of adjacent genes, but transcription of neighboring genes is surprisingly also influenced.
As a result of the multi-species analysis, one can connect species-specific chromatin regulation, TE-mediated epigenetic effects, natural selection against TEs, and genomic TE abundance that is unique to each species with sufficient power and appropriate phylogenetic resolution. The epigenetic impacts of a selfish genetic parasite on host chromatin landscapes are important in shaping genome evolution.
The transposable element (TE) is a widespread genetic parasite that copies and inserts itself across host genomes. A TE could impair the functions of the host genome if it is present and moves around. TEs cause chromosomal rearrangements through nonhomologous recombination that damage genes and functional elements.
Despite this, TEs have occupied nearly all eukaryotic genomes surveyed due to their ability to self-replicate. Eukaryotic genomes are characterized by both genomes with fewer genes but rich in repeats and genomes with many genes. Heterochromatic genomes often have fragmented TEs, rendering them incapable of replicating.
TEs of the euchromatic genome, which are interspersed with functional elements, are generally capable of replicating, making them active players in not only their evolutionary dynamics but in the function and evolution of the euchromatic genome as a whole. The difficulties associated with identifying TEs in heterochromatic regions have largely forced studies to focus on TE evolution in euchromatic genomes. It is noteworthy that the diversity of TE abundances in the euchromatic genome varies significantly across phylogenetic trees.
Image Source: https://doi.org/10.3390/ijms20143597.
Since TEs are replicative, they have been able to occupy nearly all eukaryotic genomes. Despite this, their ‘success,’ or genomic abundance, varies dramatically across the phylogenetic tree, raising important questions about the evolutionary causes and functional consequences of genomic TE abundance variations.
The majority of studies about TE abundance focus on insertions in euchromatic genomes. In heterochromatic genomes, TEs are abundant; however, they tend to be fragmented and, therefore, no longer part of the ‘life cycle’ of TEs. Despite the challenges associated with assembling TEs in repeat-rich heterochromatic sequences, the role of euchromatic TEs in determining the abundance and evolution of TEs in the heterochromatic genome remains unclear.
Euchromic TE abundance varies widely according to several hypotheses. Different species within a taxon may differ based on phylogenetic signals. The wide variability in TE abundance was also attributed to TE activity variations. Natural selection removing TEs differs between species in its strength, which is another plausible cause.
Natural selection has been investigated mainly from the perspective of population genetic parameters such as mating systems and effective population sizes. Through differences in epigenetic effects of TEs driven by species-specific host chromatin landscapes, the multi-species study tested another evolutionary mechanism for how the strength of selection removing TEs differs.
TE-mediated epigenetic effects reduce neighboring gene expression on a genomic scale, which is perhaps the most surprising finding. TE-mediated epigenetic effects increase the enrichment of repressive epigenetic marks at adjacent genes, but they are not associated with gene expression within genomes.
The same conclusion was reached by comparing homologous alleles with and without TEs showing epigenetic effects. As previously reported in several model organisms, TE-induced repressive epigenetic mark enrichment does not affect neighboring gene expression genome-wide, which is consistent with the observation that TEs downstream or upstream of genes do not predominantly lead to reduced gene expression.
There is a complex relationship between repressive epigenetic modification and gene expression, the varying sensitivity of genes to the enrichment of repressive marks, and the presence of other types of variants that also influence gene expression, which may explain the limited effects of TE-mediated H3K9me2 enrichment on gene expression. TE abundance and, accordingly, the structure and function of euchromatic genomes may be shaped by heterochromatin modulators, in addition to determining the epigenetic impacts of euchromatic TEs on flanking sequences. As a consequence of the inadvertent deleterious epigenetic effects of a widespread genetic parasite, euchromatin and heterochromatin may have evolved more interdependently than previously thought.
Article Sources: Reference Paper 1 | Reference Paper 2 | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.