A research team led by the professors at MIT and the University of Arizona has developed an experiment-theory platform for programming bacterial systems to grow into arbitrary two-dimensional target structures using genetically engineered bacteria and mathematical modeling.
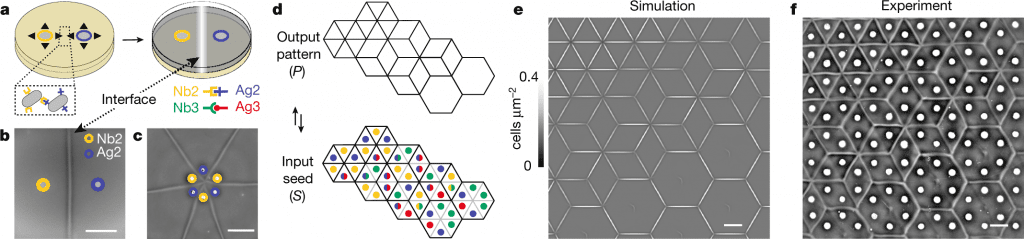
Image source: https://doi.org/10.1038/s41586-022-04944-2/
Multicellular systems, such as bacterial biofilms and human organs form interfaces (or boundaries) between different cell collectives to accomplish a wide variety of functions spatially. In the past few decades, the evolution of sufficiently descriptive genetic toolkits has likely led to the explosion of complex multicellular life and patterning. As a build-to-understand methodology for natural systems, synthetic biology aims to engineer multicellular systems for practical applications.
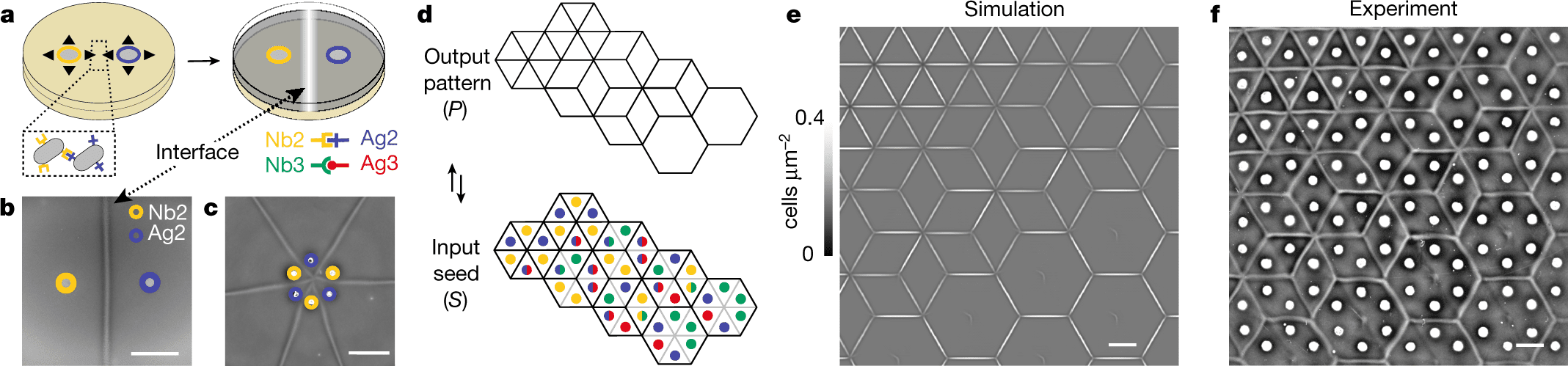
The development of synthetic cell-cell adhesion toolkits and patterning algorithms is still lacking, making it difficult to plan multicellular interface patterns. Multicellular interface patterns are engineered, modeled, and algorithmically programmed using a synthetic cell-cell adhesin logic and swarming bacteria. An adhesion-mediated analog of developmental organizers and morphogen fields is demonstrated through swarming adhesions, quantitative control of interface geometry, and adhesion-mediated interface generation.
As a result of this synthetic 4-bit adhesion logic, human-readable molecular diagnostics, fluid control on biological surfaces, and programmable self-growing materials are now possible. There is a low critical threshold for the evolution and engineering of complex multicellular systems with just four adhesins, which represent four bits of information.
Multicellular interfaces are produced by bacterial communities by cooperating and competing. Through developmental programs using genetic toolkits, organizers, and morphogen fields that are independently evolved yet similar, plants, fungi, and animals can establish tissue boundary patterns. In mathematics, such interfaces (or boundaries) can be described as tilings or tessellations.
These patterns combine generic algorithmic design principles with functional structure and aesthetic appeal in an elegant manner. Tetris video game, DNA tile self-assembly, and neuronal self-avoidance tiles are all examples of Islamic architecture exemplified by neuronal self-avoidance tilings. Multicellular systems are being engineered using synthetic biology to facilitate the development of applications such as programmable biomaterials, artificial tissues, metabolic consortiums, as well as building-to-understand methodologies for natural systems.
In spite of recent bioengineering advances, experimentally feasible algorithms and standardized modules for multicellular interface patterning remain limited. This is partly due to the fact that the design potential of cell-cell adhesion has not been explored to the same extent as the design potential of signaling between cell pairs.
It is possible to use the types of cell adhesion logic and algorithms we have introduced to engineer and investigate multicellular patterning on non-bacterial systems, such as active particles or mammalian tissues. The logic of patterning could be further complicated in engineered systems, for instance, by using sequentially established interfaces as organizers in hierarchical processes and limiting its scope by identifying system-independent or environmental inhibiting factors.
Natural systems coordinate animal morphogenesis with a differential cadherin adhesion code. Families of polymorphic adhesins are used by bacteria, amoebas, and fungi to pattern their cells spatially. Unlike chemical-based and adhesion-based mechanisms, the ‘swarming adhesion’ mechanism uncovered here may serve to distinguish between kin and foe in natural microbial communities to prevent invasion. A number of non-adhesion-based patterning mechanisms or hybrid mechanisms such as Eph–ephrin interactions that coordinate adhesions are also outlined. Furthermore, the ability to generate universal interface patterns in a plane using a combinatorial adhesion logic with only four adhesins indicates a low critical threshold for multicellular systems to evolve.
The logic for adhesion with 4 bits
The lack of interference with interface formation by the addition of a third non-complementary adhesive strain provides evidence that multistrain mixtures can be composed. Each seeding point can be manipulated to yield 16 ‘elements,’ each of which corresponds to 4 bits of information, and of which nine are actually useful. Self-interaction prevents any other combinations from being of practical interest here, but they might have applications in future patterning processes.
Two doublet elements form a visible interface only if and only if they differ. The term ‘hidden’ interface is also introduced for cases in which there are no visible interfaces, which will be conceptually helpful. The interface-formation logic was followed in all experiments that paired singlets with doublets. As a result, these orthogonal adhesins and their corresponding sets and subsets of strains are used to generate visible and hidden interfaces between cells.
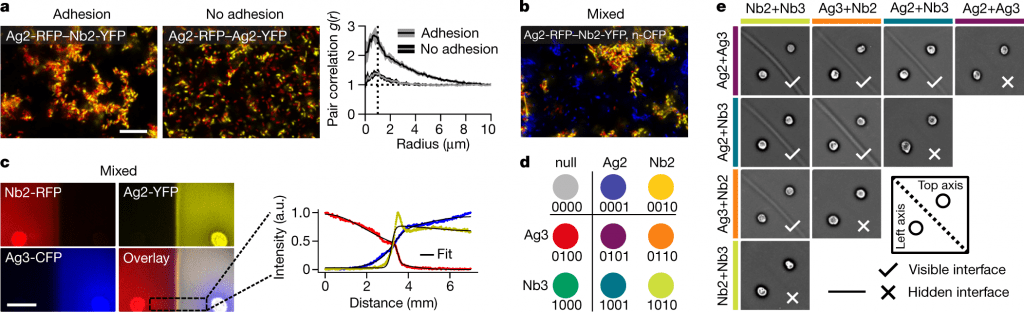
Image source: https://doi.org/10.1038/s41586-022-04944-2/
Self-growing bacterial materials: applications
The goal was to explore the potential application space of such self-growing bacterial materials by spatially patterning their properties. The basis for free-standing biomaterials can be established by cutting out patterns on thin agar sheets. At visible Nb3|Ag3 interfaces, phosphate-buffered saline (PBS) flows more slowly than at hidden Nb2|Ag3 or Nb3|Ag2 interfaces. This is due to spatial control over the wettability of the surface. Open-surface microfluidics could use this phenomenon to localize liquid droplets. By converting molecular concentrations into switchable visual interface patterns, antagonistic antigens can control interface width. A nanobody can be formed against any molecule against which a diagnostic could be manufactured without the need to purify proteins. In addition to digital displays, selective interfaces are also used to design precision biofilters and to build synthetic microbe communities with understandable interfaces.
In the future, the team is looking forward to growing three-dimensional structures and adding additional functionalities to the bacteria, such as the capability to produce specific chemicals in desired locations.
Article Source: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.