The Chinese Academy of Sciences researchers introduced a dual-plasmid editing system that enhances DNA digital storage. The study strongly encourages the use of DNA-based information technology by demonstrating a digital-to-biological information processing approach for highly effective data storage, amplification, and rewriting.
In recent years, DNA has gained increasing attention as an information storage medium. A grand challenge remains in rewriting intracellular DNA digitally in a target-specific manner since the sequences encoded in DNA are highly repetitive and have uneven guanine-cytosine contents. Using gene editing techniques, a dual-plasmid system was introduced into Escherichia coli to process this information accurately.
A high level of rewriting reliability of 94% was achieved by combining binary data containing repeat units, such as text or codebooks, with in-vivo rewriting of images. Data processing at the molecular level has been improved by introducing a new optical reporter. Over hundreds of generations, rewritten information was preserved and amplified. A biologically based approach to data storage, amplification, and rewriting is demonstrated, supporting the application of DNA-based information technology based on digital information.
The vast amount of digital information generated by the advent of the internet can be stored on a molecular scale using deoxyribonucleic acid (DNA), a DNA-based biological, genetic information carrier. In vitro enzyme reactions have not been able to provide high-precision addressing in DNA sequences, leaving DNA as a strictly writing or reading medium. Target-specific addressing and processing of digital information in living cells remain a challenge due to the kaleidoscopic variety of digital data and the uneven distribution of bases in DNA sequences.
As genome editing technology advances, new tools will be available, which will be able to repair damaged sequences, address specific target sites, and revise specific genes within living cells. These advances have inspired the development of new biotechnology for versatile in vivo rewriting of exogenous information. A major advantage of info plasmids is that digital information encoded in them can be stored and hidden stably in microbial colonies for hundreds of generations, greatly improving security during storage and transport.
Molecular biology has long used CRISPR systems to edit genes. It appears that a variety of CRISPR-associated proteins (Cas) successfully address and rewrite information-encoded DNA sequences by cleaving a target locus guided by their CRISPR RNAs (crRNAs). The protospacer-adjacent motif site limits Cas’ recognition function, and guided RNA’s secondary structure severely limits its editing efficiency, making CRISPR-Cas tools rather limited. Therefore, it is difficult to edit gene sequences arbitrarily. Additionally, information-encoded DNA sequences contain highly repeating binary codes that record a wide range of digital information, posing a large challenge for the application of CRISPR-Cas tools in DNA-based information storage because they are not compatible with endogenous DNA sequences. Thus, targeting specific DNA sequences within living cells remains an unexplored area.
Using CRISPR-Cas12a as its endonuclease, a dual-plasmid system has been developed for the storage and processing of DNA-based information. With the system, digital text, codebook, and image information were successfully stored in high density, rewritten reliably, and amplified with outstanding amplification stability. A dual-plasmid system was applied for DNA-based information storage and targeted rewriting of sequences encoded by information within living cells in this study.
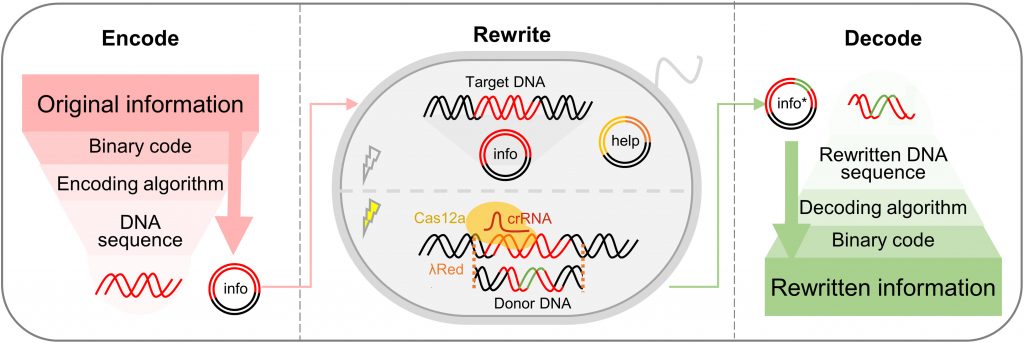
Image source: https://doi.org/10.1126/sciadv.abo7415.
As the system is compatible with various coding algorithms and does not require any addressing indices or backup sequences, it fully exploits the coding capability of DNA sequences. In the 15-ary Huffman algorithm, a compression algorithm with rotation mapping was applied to improve the coding efficiency of DNA-based information. By reducing the length of certain information containing repeats and avoiding homopolymers, the encoded DNA sequence could be reduced. By using the present systems, the coding efficiency eventually reaches 4.0 bits per nucleotide, which may be an alternative approach to improving coding efficiency.
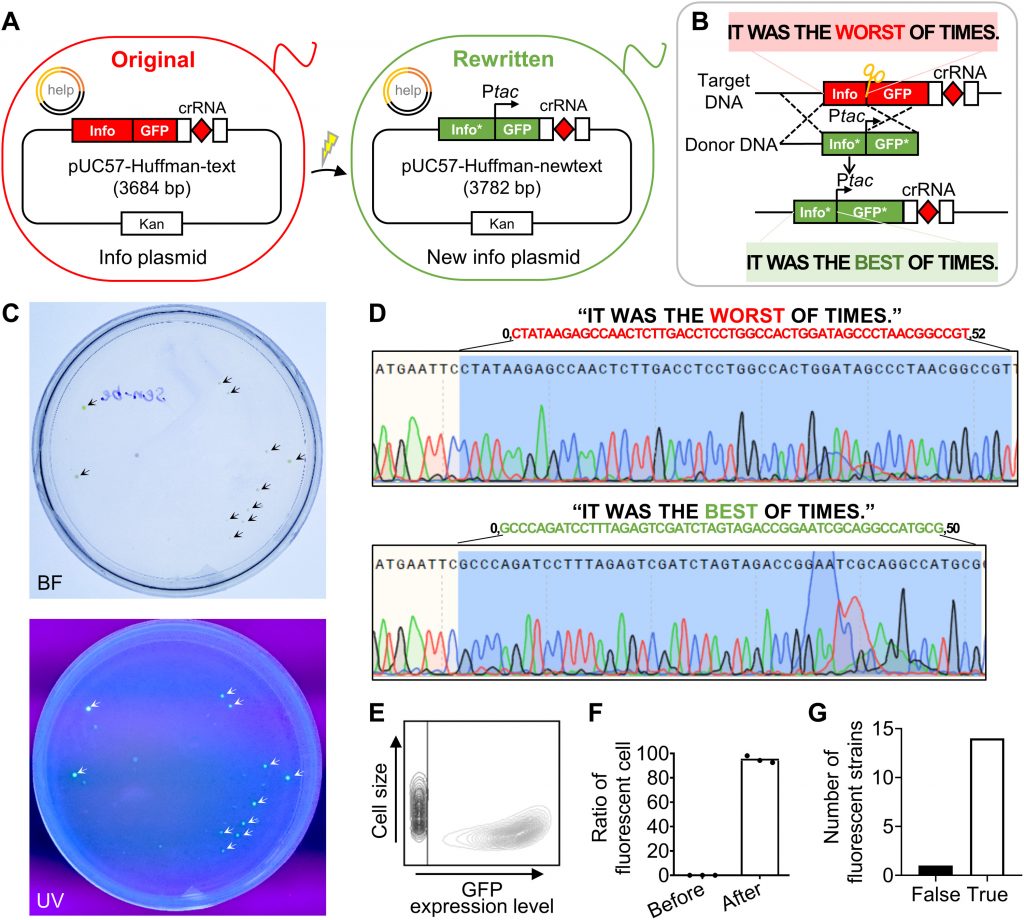
Image source: https://doi.org/10.1126/sciadv.abo7415.
The rewriting of complex information stored in exogenous DNA sequences in vivo was performed using high specificity between complementary pairs of nucleic acid molecules. High rewriting reliability of up to 94% was achieved by optimizing the crRNA sequence, making the information rewriting tool highly adaptable to complex information.
In living cells, the dual-plasmid CRISPR-Cas12a system was used to precisely target and reliably rewrite digital information stored in exogenous DNA sequences. In a digital-to-biologic system, DNA, a molecular-level information carrier, can be treated just like physical memory with targeted specific access and information editing.
In addition, the system allows living cells to process digital information flexibly and dynamically. The size of the host genome and the amount of DNA inserted limit the storage capacity of this dual-plasmid system. The development of plasmids such as yeast artificial chromosomes and the use of plasmids in a living host with a larger genome would further pave the way for practical applications regarding big data storage.
It could also be combined with multiple functional elements, including silica biomineralization peptides and light-inducible transcription factors, to provide long-term data storage and flexible information processing, respectively. A living cell information storage array could also be built using microfluidic technology. Chemical biology has infinite potential applications across various disciplines and expands the platform for DNA-based information storage.
Article Source: Liu, Y., Ren, Y., Li, J., Wang, F., Wang, F., Ma, C., Chen, D., Jiang, X., Fan, C., Zhang, H., & Liu, K. In vivo processing of digital information molecularly with targeted specificity and robust reliability. Science Advances, 8(31), eabo7415. (2022) https://doi.org/10.1126/sciadv.abo7415.
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Srishti Sharma is a consulting Scientific Content Writing Intern at CBIRT. She's currently pursuing M. Tech in Biotechnology from Jaypee Institute of Information Technology. Aspiring researcher, passionate and curious about exploring new scientific methods and scientific writing.












