With a thorough potential of the Ribonucleic Acids (RNA) in cellular processes, the molecule is deemed more than just being transitionary and has emerged as a master regulator of cellular processes. The regulatory functions to code proteins and untranslated regions in mRNA significantly contribute towards post-translational modifications. A review published in Nature Reviews Genetics gives biological insights into understanding the RNA ensemble and its potential using next-generation sequencing technology.
Introduction
The extensivity of the human genome lies in the genomic material in the form of DNA and RNA. The earlier research regarded the biological processes with RNA as a transitionary state for transcription to translation. However, RNA is also a crucial regulator of almost all cellular processes, and the complex structural organizations adopted by the RNA molecules are thought to often be pivotal for their functions and regulation in living cells. Several studies have inculcated the research to understand the dynamics of the RNA complex organization-The RNA structurome.

Image Source: https://doi.org/10.1038/s41576-022-00546-w
The RNA structurome identifies itself as a stable entity enabling researchers to dive deep into the intricate networking of alternate intramolecular and intermolecular interactions. Being the master regulators and existing with expanded repertoires such as microRNAs and snRNAs can mediate the functions such as gene expression, transcription, and post-transcriptional modifications. Hence, they deem it important to elucidate the function mechanism. The current reviewers- Danny Incarnato from the University of Groningen and Robert C. Spitale from the University of California, Irvine in California, USA, have described the latest advancements in NGS technology to determine the RNA structure based on large-scale transcriptomics in biological cells.
Reviewers have extensively discussed the structural heterogeneity of the RNA molecules and networks and how high throughput sequencing and computational methods can assist in mapping them. The review has highlighted the ability of the RNA functional units to engage and regulate intracellular and intercellular communication on a cellular level. Finally, the reviewers have also addressed the open-ended questions with the possibility of challenges associated with their role in therapies. Understanding the functionality of RNA structure also provides directional study into emerging novel therapeutics such as personalized RNA therapy.
High throughput analysis for RNA structure is performed in sequential steps using complementary approaches such as chemical and biochemical methods that have been cultivated to identify the contribution of RNA structure in mediating the RNA–protein interactions and binding. The approaches intersected with NGS technologies can provide data on the structural organization, interaction, and substitution between the different RNA molecules. The sequential steps include:
- Structure probing of individual nucleotides: More commonly termed as chemical probing, is performed specifically on functional groups. The chemical approach is extensively used to determine the structure of RNA molecules. Dimethyl Sulfate (DMS) is a commonly managed reagent to measure the base-pairing, similarly, potent SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) probes can also be developed to quantify the RNA structural flexibility.
- Mapping intra-molecular and inter-molecular RNA–RNA interactions: Several indirect methodologies have been constructed for the group and outline these RNA-RNA interactions. The direct methods include two types of chemical probes: one being those that cross-link the base-paired regions and the other that cross-link structurally closes RNA functional groups. The experiments adjoined with HTS enable using tools such as psoralen analysis of RNA interactions and structures (PARIS) and mapping RNA interactome in vivo (MARIO).
- RNA structure characterization of RNA–protein interconnection: Understanding the interface of RNA-protein interactions will help elucidate the overall structural characterization in-vitro setting. RNA–protein interaction can be evaluated using indirect methods by measuring changes in probe reactivity between the free RNA and RNA, i.e., protein-bounded. A few of the approaches include the footprinting SHAPE (fSHAPE) uses differential SHAPE examining in vivo, and ex vivo deproteinized conditions to identify associations.
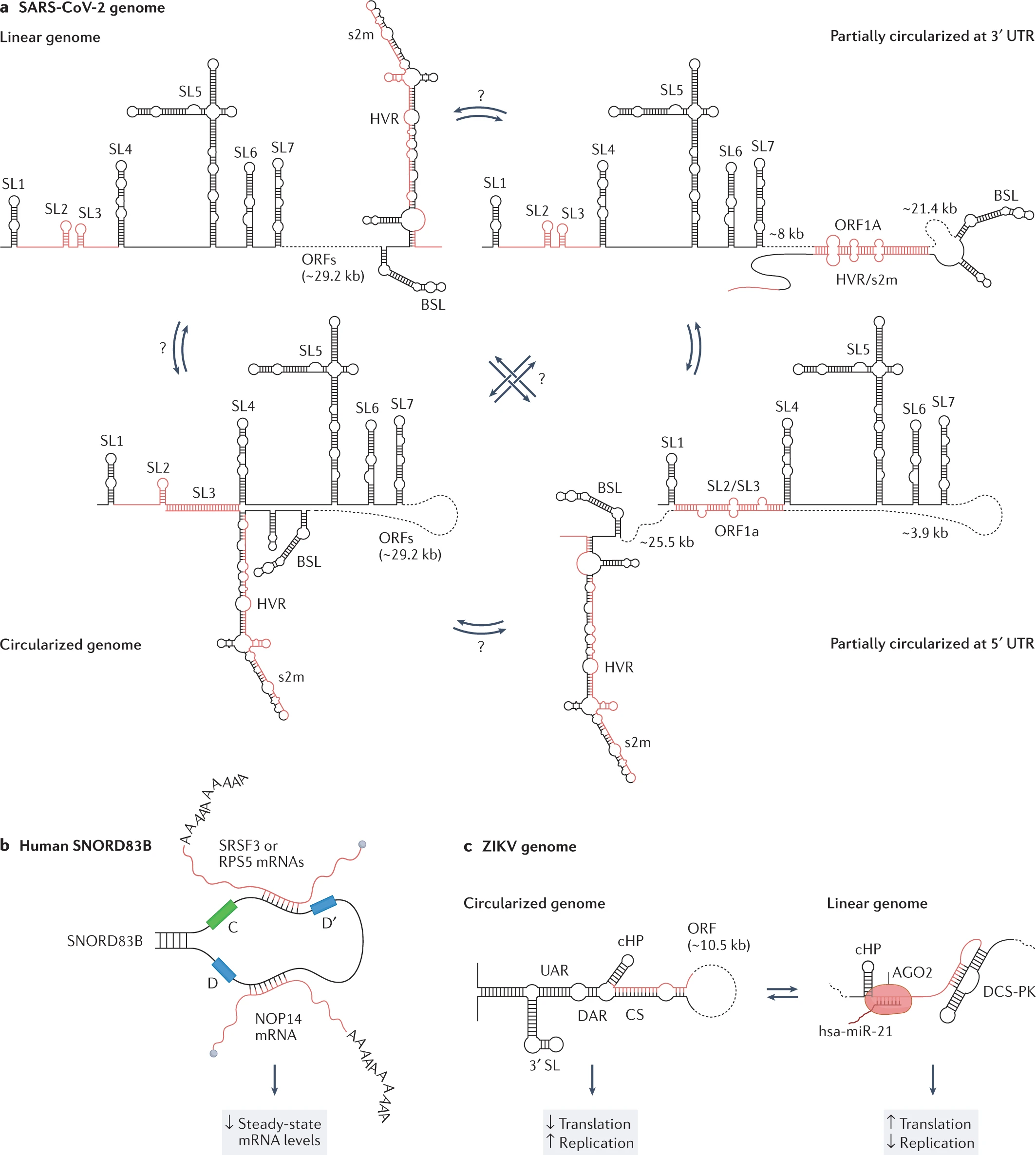
RNAs have promptly gained momentum in research associated with drugs and therapies. Hence, the reviewers believe dedicated information on the ‘structurome’ is still very restricted. Identifying and developing the architecture for RNA-RNA interactions can provide a potential role in therapeutics. Mapping the RNA bipartite using PARIS in human and mouse cells has provided data for 30%-40% relative bipartite interactions. The long-term interactions into cellular dynamics can be proven more beneficial than the short-sighted interactions established in ZIKV and DENV genomes.
Considering the RNA heterogeneity, few attempts have been made to ensemble RNA structure in the living biological cell. The experimental and computational methods for RNA ensemble of the signal and image processing for HIV-1 viral replication were set to regulate the RNA structural switches. The Rev protein recognition element (RRE) is crucial to identify the major and minor conformations. Similarly, two respective independent studies also reported the ensemble of the signal and image processing analysis of the SARS-CoV-2 genome, probed by DMS-MaPseq under either the in vivo or in vitro conditions.
Final Thoughts
The reviewers have extensively elucidated the complexity of RNA structurome. The review has provided a crude outline of RNA structure ensembles since they can detect the major structural realignments. The outlining work has highlighted various approaches to identify the complex networking and RNA-RNA and RNA-RNA protein interactions. The methodology of these approaches has been extended from traditional methods to high-throughput sequencing technology and further advances the sensitivity of these methods for an applicative role in therapeutics and RNA-based drug technology.
Article Sources: Reference Paper | Reference Article
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Mahi Sharma is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She is a postgraduate with a Master's in Immunology degree from Amity University, Noida. She has interned at CSIR-Institute of Microbial Technology, working on human and environmental microbiomes. She actively promotes research on microcosmos and science communication through her personal blog.