Recent research conducted by the research group of Savas Tay at the University of Chicago, Illinois, USA, introduced a deep learning platform, OrganoID to analyze and interpret the organoid models for personalized drug screening. The team has interpreted results and validated the platform using pancreatic cancer organoid images, revolutionizing biomedicine.
Introduction
Organoid research has risen up drastically in biomedical research due to its expandable potential in understanding disease in 3D cellular form. Throughout the years, the research has been directed by using murine models for studying diseases and screening for potential therapeutics. Whilst, organoids closely enumerate the heterogeneity on a cellular level, structural morphology, and functions of a variety of tissues with organ specificity that provides an advantage over using the traditional single-cell cultures.

Image Source: https://doi.org/10.1371/journal.pcbi.1010584
Since organoids appear as better models, the limitations associated often result in major challenges for the experiments associated with the organoid experiments due to the analysis and measurement of drug responses. Considering the large scale of microscopy images that are analyzed for comparison and extract information, image analysis is deemed as difficult. Various technological advancements and optimizations have surfaced throughout the years for image analysis of organoids across all the focal points and tissue heterogeneity in the organ. Some of them include Zhou Z et al. and Mead BE et al. platforms relying on pre-image and pre-tuning of the image based on the parameters of brightfield image analysis.
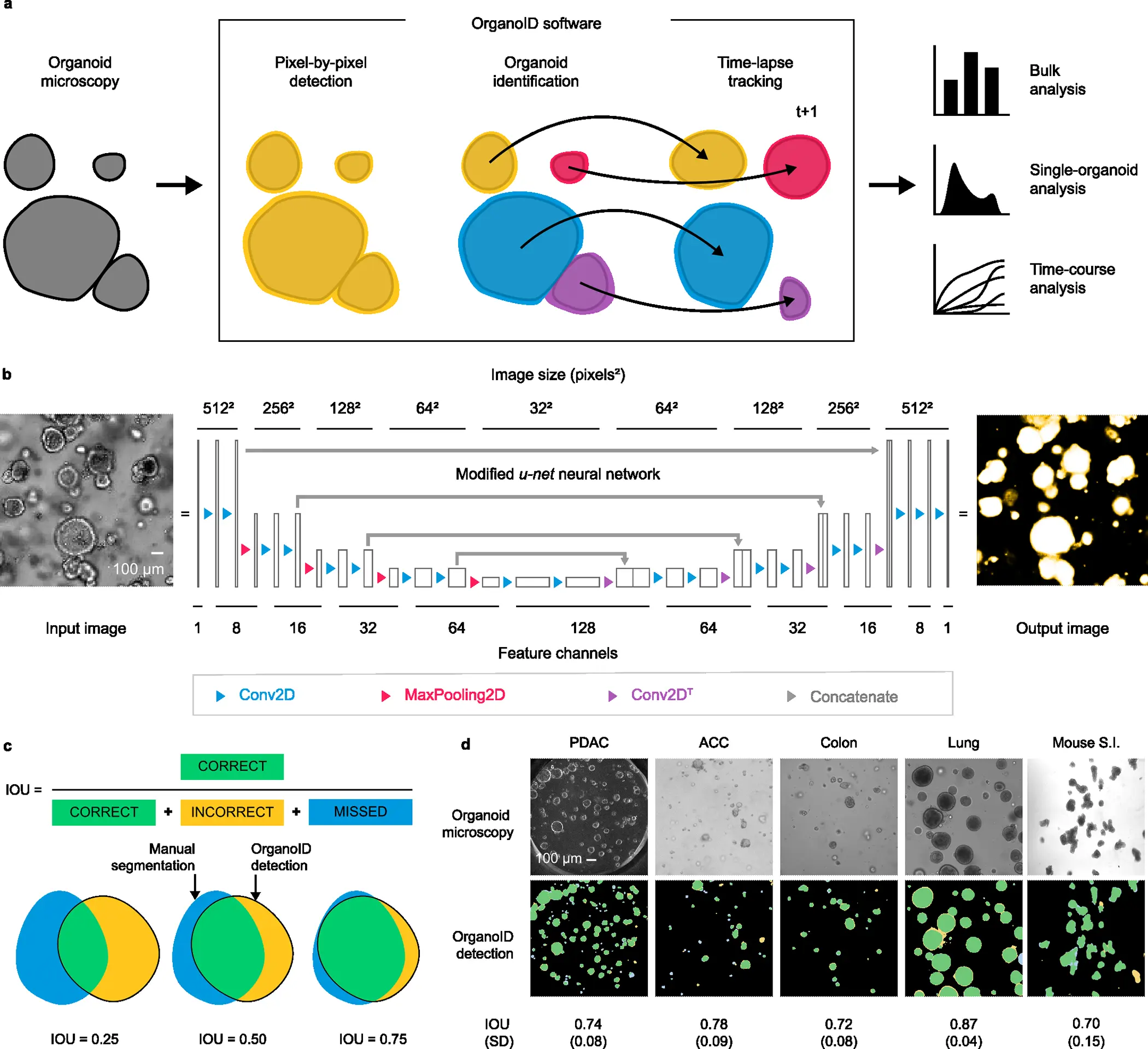
Researchers from The University of Chicago, Illinois, USA, have overcome these limitations by introducing the deep learning platform OrganoID, a robust platform for image analysis that efficiently and automatically recognizes and labels the images and tracks single organoids, pixel-by-pixel in experiments set up in brightfield and in phase-contrast microscopy. The team has trained the platform using the images of the organoid model for pancreatic cancer and validated using the bulk of images associated with pancreatic, colon, and lung organoids. The team has provided its applicative responses as most reasonable due to its ability to not only identify the images accurately but also advance in data-driven applications such as drug exploration and personalization.
The OrganoID platform consists of three major components that allow them to have consistent and accurate segments. The three components are:
- A convolutional neural network that utilizes microscopy images for the detection of organoids.
- An identification module that resolves and contours subsequently to label the individual organoids.
- A tracking module that follows the identified organoids in time-lapse imaging throughout the experiments.
The overall design and implementation were based on first developing a convolutional neural network. A deep learning-based pipeline was constructed for image analysis, and the platform appoints a convolutional neural network to reconstruct the microscopy images into organoid probability images, brightening the overall images and present the organoid at a provided pixel. The network structure for the platform was derived from the extensively successful u-net approach for image segmentation. The platform utilizes and regards the u-net structure.
In lieu of the training of the platform, an original dataset consisting of a total of 66 brightfield and phase-contrast microscopy images of the organoids. These images were manually segmented to construct ground truth images, black-and-white, for the training of the network using 52 image pairs and 14 image pairs for the validation. Approximately 5-50 images from the samples of two patients of human pancreatic ductal adenocarcinoma (PDAC) were utilized. The network orientation and segmentation for training the dataset were processed using the Augmentor Python package.
Post constructing the convolutional model for individual organoids, the identification was laid by grouping the analysis pixel by pixel. In order to correspond to different clusters of organoids, the research team constructed an organoid separation pipeline that uses the raw network to predict the image and group pixels into single-organoid clusters. Surpassing the conventional image segmentation method using a neural network, the approach was more effective in determining the information and associated strength of the predictions.
The team employed a Canny edge detector to identify the image contours that are able to modify the age as such: 1) Firstly, it computes the partial derivative of pixel intensity to identify areas of high contrast. 2) Secondly, it also blurs the image to smooth out the noisy regions. 3) Thirdly, it employs a hysteresis-based threshold to identify strong edges locally. The resulting identification from the pipeline provides an output of a labeled image, wherein the pixels representing an individual organoid are all set to a distinguished number for the ID, which can also be applied for single-organoid analysis.
Finally, the automation is applied to the database of organoid images, each representing a unique ID. The researchers have developed a tracking module for a longitudinal analysis of the single organoids using time-lapse experiments for imaging. The overall changes in various landscapes of each individual organoid, such as the shape and size, can be thoroughly followed and measured. The tracking algorithm of the deep-learning platform built a cost-assignment matrix for each individual image in a sequential time-lapse manner.
Since the trained model was primarily based on PDAC organoids, the team has also extended the study to evaluate its capacity to generalize to the organoids derived from non-PDAC tissues, such as the epithelial cells from the lungs and colon. The OrganoID network passed the benchmark for all the images of non-PDAC, with a mean IOU of 0.79 (SD = 0.096). The results support the potential of the model’s neural network in generalizing the images from various tissues without any specificity tuning for identification.
The extended work for drug screening is also feasible as the researchers have deemed that the changes in organoid physiology and structure can indicate the key responses for both of them being phenotypic and state transitions. OrganoID is believed to automatically outline the morphology of the organoids individually across the gemcitabine dose-response experiment resulting in organoid circularity, solidity, and eccentricity, displaying the sigmoidal dose responses of the drug and also determining the endpoint EC50 to be 28.47 nM, 27.05 nM, and 18.08 nM.
Final Thoughts
Organoids have revolutionized biomedical research, and the improved deep learning platform adds to the learning associated with complex 3D models. The OrganoID bridges the gap and automates the process of accurate pixel-by-pixel organoid identification and tracking almost in real time using time-lapse experiments. Since it is trained on broadly available microscopic images, it allows better optical configurations. The platform can subsequently also identify the morphological features of organoids without using traditional methods of cytotoxic dyes. While certain limitations can help optimize the model, such as the platform can appropriately identify the contours of organoids in physical contact while it cannot distinguish organoids that often overlap across the focal plane. Overall, it also provides a potential that can be addressed for deep-learning screening of drugs and personalization.
Article Sources: Reference Paper | OrganoID Platform | Dataset
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Mahi Sharma is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She is a postgraduate with a Master's in Immunology degree from Amity University, Noida. She has interned at CSIR-Institute of Microbial Technology, working on human and environmental microbiomes. She actively promotes research on microcosmos and science communication through her personal blog.