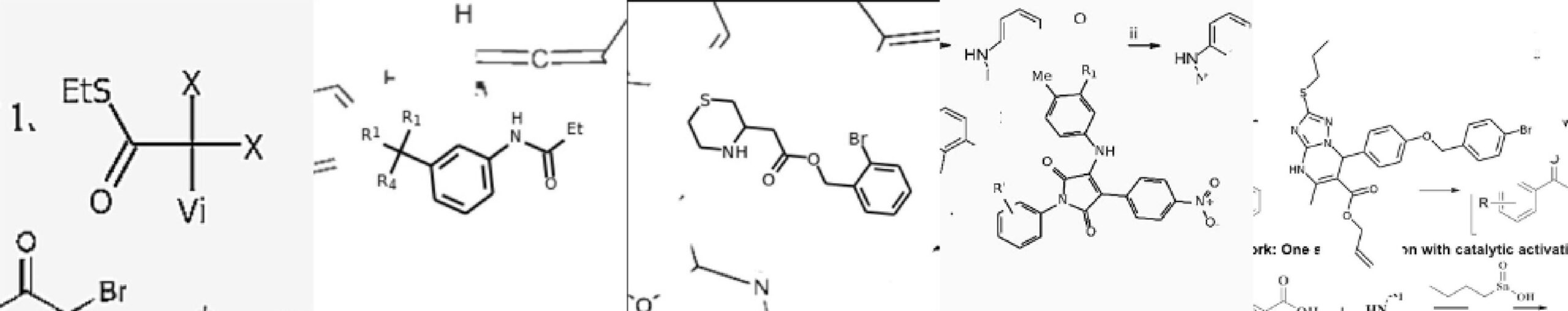
The researchers from Syntelly — a startup that originated at Skoltech — Lomonosov Moscow State University, and Sirius University present a Transformer-based artificial neural network that can turn images of organic structures into molecular templates.
The model is capable of automatically detecting chemical formulae on research paper scans. The findings were published in Chemistry-Methods, that is a European Chemical Society journal.
The age of artificial intelligence has arrived for humanity. Deep learning’s emergence in numerous scientific and technological fields encourages the development of AI-based information retrieval systems. Modern deep learning technologies, which typically demand enormous volumes of qualitative data for neural network training, will also be beneficial in the field of chemistry.
The good news is that chemical data holds up well over time. Even though a molecule was first synthesized more than a century ago, knowledge regarding its structure, characteristics, and methods of synthesis is still useful today. Even in this day of ubiquitous digitization, an organic chemist may consult an original journal paper or thesis from a library collection — like, from the early twentieth century in German — for information on a poorly researched molecule.
The sad thing about this is that there is no universally agreed method of expressing chemical formulas. Chemists employ a variety of shorthand notation strategies to represent common chemical groupings. The possible stand-ins for a tert-butyl group, for example, include “tBu,” “t-Bu,” and “tert-Bu.” To make matters worse, scientists frequently employ a single template with many “placeholders” (R1, R2, etc.) to refer to a large number of identical compounds, but those placeholder symbols might be described anywhere: in the figure itself, in the article’s main text, or in supplements. Not to mention that drawing styles fluctuate from journal to journal and evolve with time, scientists’ personal preferences shift, and norms change. As a result, even a seasoned scientist might become perplexed while attempting to solve a “mystery” discovered in a magazine article. The problem looks intractable to a computer algorithm.
Image Source: Image2SMILES: Transformer-Based Molecular Optical Recognition Engine.
The automated extraction of chemical information relies heavily on the optical recognition of organic structures. An optical recognition system may be built using one of two methods: first, a rules-based technique in which optical primitive detectors (character, lines, and edges detectors) are used in conjunction with a set of pre-defined rules that characterize typical journal drawing styles and second, a deep neural network-based technique that is entirely data-driven.
Machine learning relies heavily on data. However, there are no open-access datasets containing annotated objects on chemical articles. Building a data-generative model is the only approach to getting a huge dataset. This model should be able to reproduce a wide range of real-life paper drawings. We feel that Transformer cannot be adequately trained without a robust data augmentation method.
This method is unique in that it focuses heavily on data production schemes and has the ability to handle organic structures and molecular templates, making it suitable to use on actual data.
The researchers had already tackled comparable challenges with Transformer, a neural network first developed by Google for machine translation, as they approached it. The researchers employed this sophisticated technology to transform the picture of a molecule or a molecular template to its written representation rather than translating words across languages. ‘Functional-Group-SMILES’ is a name for such a representation.
The neural network was capable of learning practically anything, much to the researchers’ astonishment, as long as the appropriate representation style was included in the training data. Transformer, on the other hand, needs tens of millions of instances to train on, and manually gathering that many chemical formulae from research publications are difficult. Instead, the scientists used a different method and developed a data generator that generates molecular template instances by merging randomly picked molecule fragments and portrayal styles.
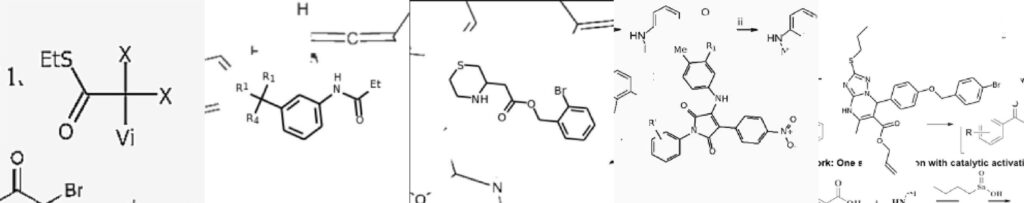
Image Source: Image2SMILES: Transformer-Based Molecular Optical Recognition Engine.
The principal investigator of the study Sergey Sosnin, the CEO of Syntelly, a startup founded at Skoltech, said – “Our study is a good demonstration of the ongoing paradigm shift in the optical recognition of chemical structures. While prior research focused on molecular structure recognition per se, now that we have the unique capacities of Transformer and similar networks, we can instead dedicate ourselves to creating artificial sample generators that would imitate most of the existing styles of molecular template depiction. Our algorithm combines molecules, functional groups, fonts, styles, even printing defects, it introduces bits of additional molecules, abstract fragments, etc. Even a chemist has a hard time telling if the molecule came straight out of a real paper or from the generator.”
The study’s authors think that their technique will be a significant step toward developing an artificial intelligence system capable of “reading” and “comprehending” research articles to the same level as a highly-skilled chemist.
Story Source: Khokhlov, I., Krasnov, L., Fedorov, M. V., & Sosnin, S. (2022). Image2SMILES: Transformer‐Based Molecular Optical Recognition Engine. Chemistry‐Methods, 2(1), e202100069. https://doi.org/10.1002/cmtd.202100069
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.