Drug repurposing has been strategically used to modulate the usage of pre-existing drugs. The virtual screening tool DrugRep facilitates computational screening using both receptor and ligand-based methods. Being freely available, it provides the unstinted potential for repurposing and investing in current pharmacology.
Historically, introducing novel drugs bears challenges in terms of exploring and introducing them for healthcare purposes. Throughout the years, novel drug development demands a prolonged timeline alongside tried and failed clinical trials before launching them into the scalable market of pharmaceuticals. In order to expand the potential of existing drugs in different diseases, drug repurposing was introduced to repurpose drugs by identifying new drug targets computationally. The sudden collapse of the healthcare industry during the COVID-19 pandemic resurged drug repurposing using CADD (Computational Aided Drug Design).
Addressing the advantages of repurposing, a team of researchers from Sichuan University, Chengdu, China, constructed an automated virtual screening tool DrugRep. With its easy-to-use interface, the tool facilitates drug screening for both receptor-based and ligand-based methods. Considering the successful application of in silico screening methods, such as the Gleevec drug prescribed for chronic myeloid leukemia, it was repurposed for malignant gastrointestinal tumors, and Aspirin was repurposed effectively for colorectal cancers and heart conditions.
The virtual screening methods are classified as receptor-based or structure-based screening as it uses affinity for chemical compounds using molecular docking algorithms based on protein conformations. Several tools for structure-based screening have surfaced, such as SwissDock, Pocket, CavityPlus, etc. Unlike receptor-based screening, the tool also utilizes ligand-based methods using screening of the 3D structure of the receptor. Some of the methods employed include Open Babel PF2 for identifying new compounds based on similar properties of active compounds. While tools with such screening are iScreen, SwissSimilarity, and Ligity.
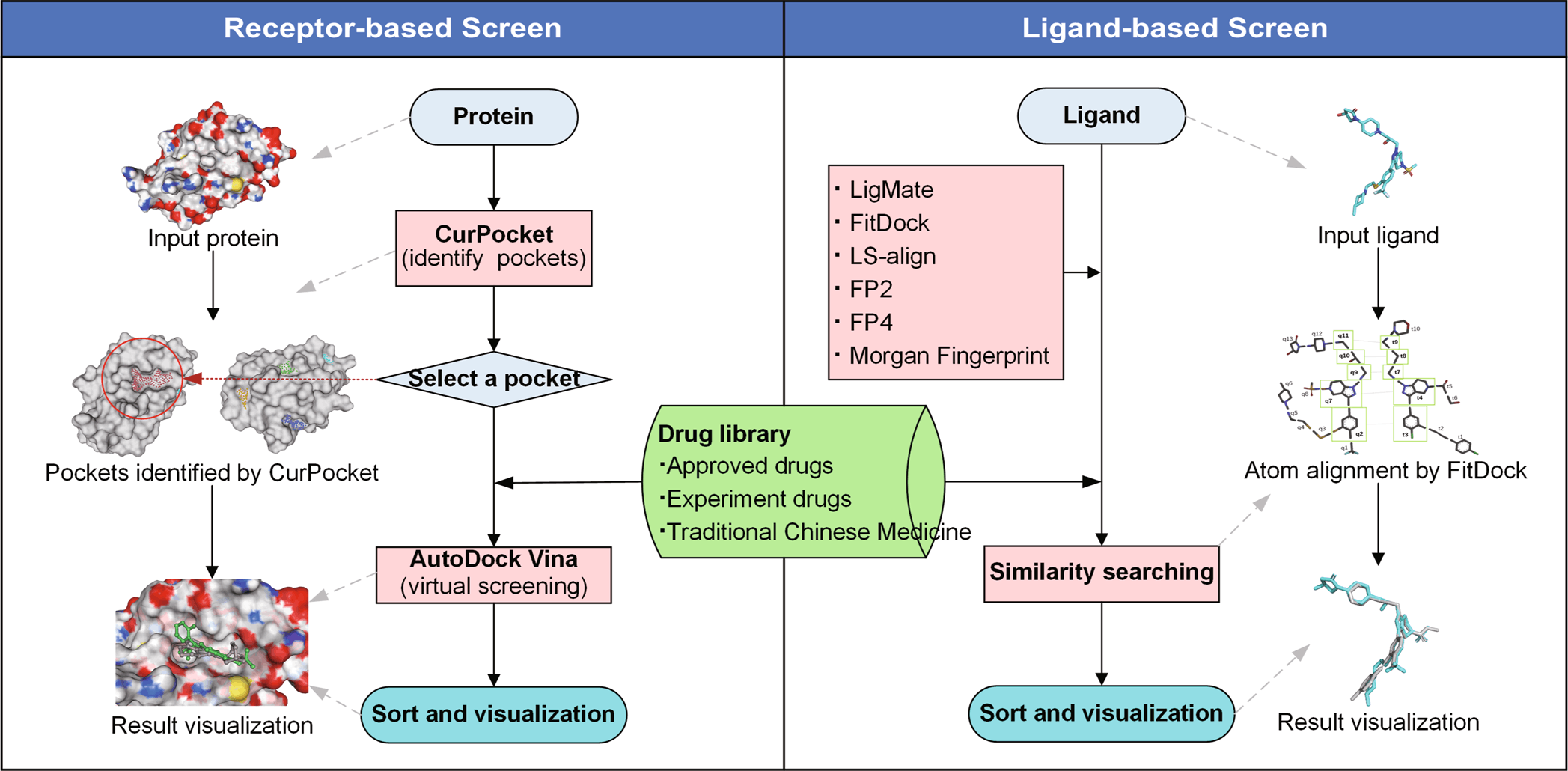
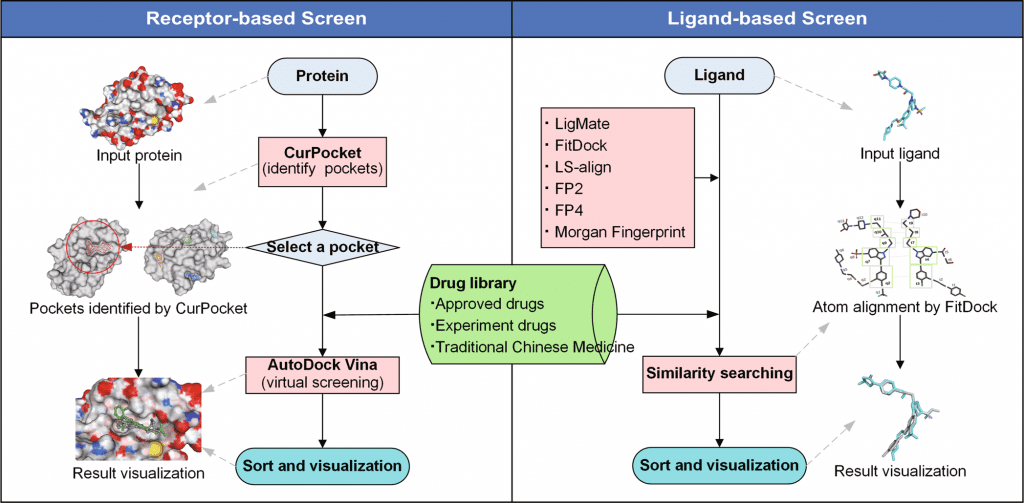
JH Gan et al. and team constructed the workflow of DrugRep, using three-drug libraries- approved drug library, experimental drug library, and traditional Chinese medicine library- performed RBS (Receptor-based screening) and LBS (Ligand-based screening). Several in-house integrations were also presented in the tool, such as CurPocket- structure-based cavity detection for RBS using calculated curvature factors and LigMate- ligand-based virtual screening.
Since the web interface is built using HTML5, CSS3, Javascript, and PHP libraries, it provides easy to use consumer-based interface. In addition to structure-based detection, RBS also employs a co-crystallized ligand from PDBbind. The three-drug libraries, approved drug library with a dataset of 2315 drugs, experimental drug library with 5935 drugs dataset, and traditional Chinese medicine library with over 2390 drug datasets were included in the tool.
The benchmark test was established based on the established virtual dataset with 40 protein targets and 98,266 active compounds-DUD. Similarly, enhanced datasets of DUD, DUD-E, and MUV (Maximum Unbiased Validation) were used for benchmark tests. The performance of screened datasets was shown by Receiver operating characteristic (ROC) curve. The performance of RBS achieved an average of 0.69 on AUC values which were comparatively higher than other docking box results such as Durrant (0.66) and DrugRep-xtal (0.74). Similarly, the average of EF1 and SAHH was notably higher for DrugRep (19.79, 26.84), outperforming other tools. Considering the results, the team displayed high accuracy and repurposing without constricting parameters.
The performance of LBS was evaluated upon calculating the AUC AND EF1 value for selected active compounds on each target on the MUV dataset. The procedure was done in 100 replicates in order to reduce the randomness impact. The histogram data showed LigMate’s higher targets as compared to control methods. The enrichment test of complementary methods (LigMate and FitDock) of DegRep showed better performance compared to other controls (MACCS, Morgan fingerprint).
Researchers used case studies to analyze the overall performance using the nonsteroidal anti-inflammatory drug (COX-2 inhibitor). COX-2 inhibitors are known to have potential effects on heart-related complications during clinical trials. Hence, a simple process to use a virtual screening server is used as follows for COX-2 screening for receptor-based screening:
1) Upload the target protein of interest in PDB format.
2) Detecting and selecting the pocket to run screening.
3) Submitting the task on the server and viewing the results of RBS.
The best 30-scored drugs, such as bendazac, meclofenamic acid, and phenylbutazone associated with COX-2, would be identified. The similarity ligand score for identified drug targets can also be assessed for ligand-based screening. Hence, the tool DrugRep can be used to not only identify the potential targets and binding patterns.
Final Thoughts
Considering the overall contribution of the economy, manpower, animal screening, and human clinical trials that is input into drug discovery, it is indeed challenging and costly. The improvisations in computational-aided drug design facilitate the screening and repurposing of existing drugs. The scientist’s effort in constructing a virtual screening server DrugRep provides advantages in benchmark tests to other tools while the team acknowledges limitations. Some restrictions are false positive predictions due to binding conformations or low-affinity active compounds. Overall, DrugRep outperforms other similar tools and can be simulated for better usage by optimizing pipelines and stabilizing the computation.
Article Source: Reference Paper | Web site: DrugRep
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Mahi Sharma is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She is a postgraduate with a Master's in Immunology degree from Amity University, Noida. She has interned at CSIR-Institute of Microbial Technology, working on human and environmental microbiomes. She actively promotes research on microcosmos and science communication through her personal blog.