An epidemic of monkeypox occurred in May 2023 that led to a disease named mpox displaying symptoms such as fever, enlarged lymph nodes, and rash. This ailment is usually mild and self-limited, but severe cases can be harrowing and may cause lifetime scarring. In response to the outbreak, researchers from Mount Sinai and the Carlos III Health Institute (ICI) in Madrid, Spain, worked together to investigate the genetic makeup of the monkeypox virus (MPXV). They mainly studied subclade IIb strains of the virus, helping them to understand the convolutions in viral genes that influence virus behavior, thereby offering strategies for intervention.
Understanding Genomic Accordion
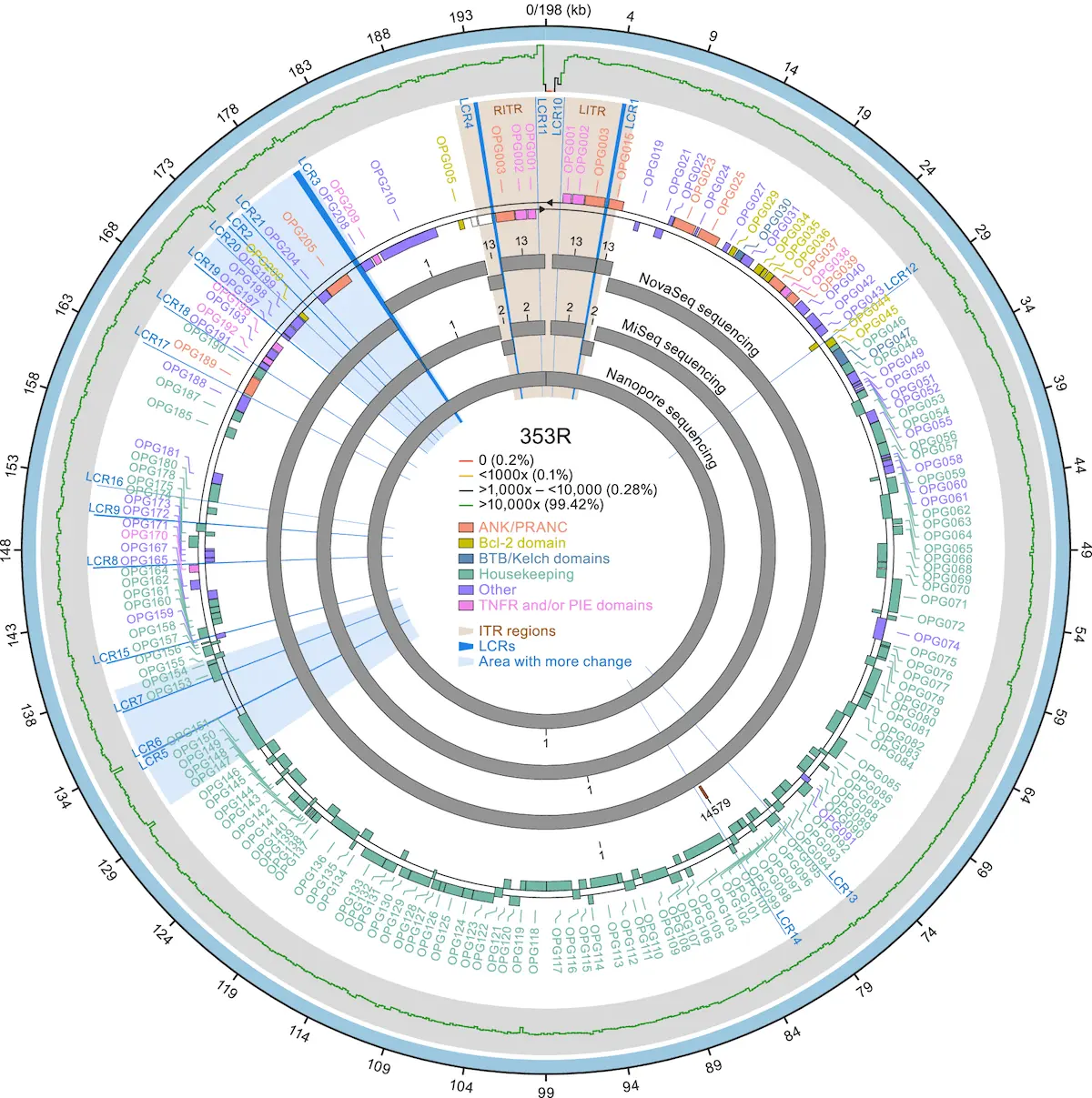
The concept of “genomic accordion” refers to periodic expansions and contractions within the genome of monkeypox virus (MPXV), especially in low-complexity regions (LCRs). Adaptive evolution for this virus depends on these variants, which are drivers of gene expression changes or modifications, including short tandem repeats.
Contrarily, researchers believe that differences in LCRs rather than single-nucleotide polymorphisms (SNPs) may explain the distinct epidemiology of subclade IIb MPXV strains. MPXV’s genetic pathways that underlie its unique transmission dynamics and pathogenicity have been illuminated through precise LCR resolution and repeat length analysis. They also fill us in on viral evolution and adaptability, emphasizing the importance of LCRs and aiding our understanding of the genomic dynamics of MPXV. The concepts behind this view are embodied in the idea of a “genomic accordion” that helps open our eyes to how viruses such as MPXV cope with different environments to guide further investigations and public health-oriented interventions.
Approaches Adopted
Swabs from the patient’s vesicular lesions were taken and analyzed at CNM (Centro Nacional de Microbiología) to address this issue. Nucleic acids were extracted from these samples using either the QIAamp MinElute Virus Spin DNA Kit or the QIAamp Viral RNA Mini Kit. Subsequently, these samples were then inactivated in a BSL-3 facility following BSL-2 guidelines. Several specific diagnostic techniques were used to confirm the diagnosis of MPXV, while others confirmed the presence of the virus.
Customized R script implemented generation of allele frequency graphs and quality control analyses for modified STRsearch, which was employed to assess low-complexity regions (LCRs) within populations using intra-host and inter-host variation tools.
LCRs and SNPs with inter-sample variability were subjected to genetic distance calculations using pairwise Euclidean distances. The phylogenetic analysis process employed Snippy for variant calling and SNP matrix creation. In IQ-Tree analysis, 1000 bootstraps were subsequently applied.
Implications
The implications of these findings are far-reaching:
Viral Evolution and Adaptability: Exploring the diversity of short tandem repeats (STRs) within low-complexity regions (LCRs) of the Monkeypox Virus (MPXV) genome can give insight into the genetic processes underlying viral evolution and adaptability. This information would be helpful in predicting future changes in the disease over time.
Therapeutic Targets: The discovery of critical transcriptional regulatory elements such as LCRs and STRs that control gene expression/disease phenotype may lead to novel therapeutic targets. It is possible to develop improved therapies or vaccines against MPXV by focusing on these areas.
Public Health Measures: Genetic analysis of MPXVs helps shape public health policies as well as epidemic response strategies. Knowing how the virus evolves genetically allows appropriate measures customized for containment and prevention of outbreaks.
Concluding Remarks
Overall, this work is a big step towards unraveling the genetic secrets of the Monkeypox Virus. The careful investigation of viral genes, LCR variability, and genomic analysis sets the door for future studies targeted at treating developing infectious illnesses and comprehending the complex principles of viral evolution.
To get more insights, read the Reference Paper and Reference Article, and the codes used in this study are available on GitHub.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.