Molecular biology scientists have been captivated by the complex way RNA folds. A recent study by Beihang and Nanjing University researchers in China introduces the RiboDiffusion model. This new method looks at predicting protein structure at the atomic level and is more advanced than existing methods like StructGNN and GVP-GNN.
RiboDiffusion: A Game Changer
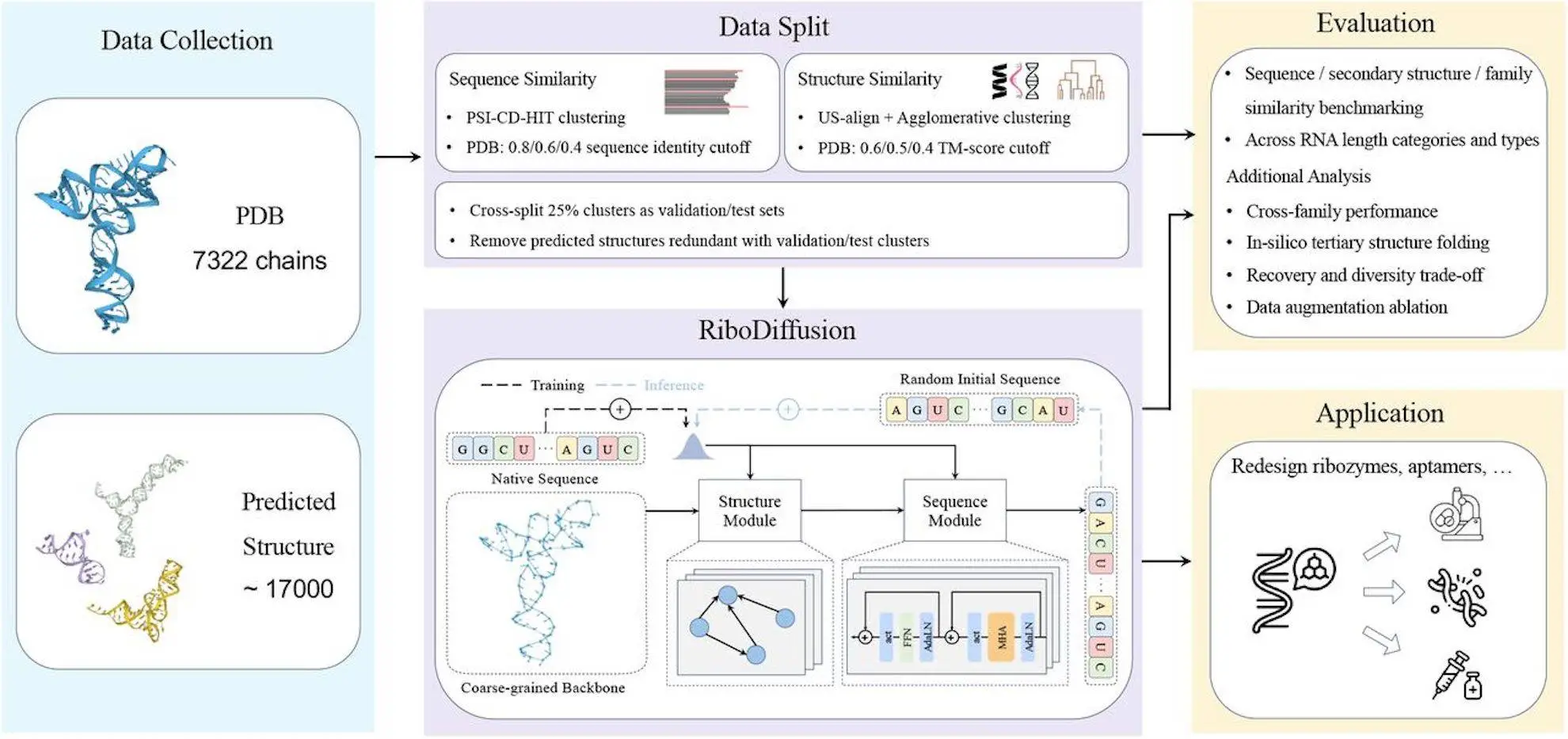
Designing RNA has become important for making synthetic biology and treatments that work in specific ways. One challenge is making sequences that fold into desired structures, which is called the inverse RNA folding issue. Initial methods focused on simple structures, but now we can use deep learning and data to map sequences to structures.
This study looks at RiboDiffusion, a new method for generating RNA structures based on tertiary structures. It can make sequences following geometric rules. The model has structure and sequence parts, which are trained using data from PDB and predicted structures. RiboDiffusion is better than other machine learning methods at finding sequences and structures for different kinds and lengths of RNA, making it suitable for designing RNA under specific rules.
Methodology: Decoding the RNA Folding Puzzle
RiboDiffusion is a deep generative model for RNA inverse folding that relies on fixed 3D backbones. It uses diffusion models, which have proven successful in various data distribution learning tasks, to perturb data with noise and learn the sequence distribution conditional on RNA backbone structures.
Inverse folding is the exploration of sequences that fold into a specified structure, which is represented by fixed RNA backbone structures. RiboDiffusion addresses this by learning the conditional distribution of sequences with fixed backbone architecture. It uses a forward diffusion technique to gradually damage data with noise, which is then reversed to yield the required sequences. This approach entails defining a scoring function to guide the reverse diffusion process.
RiboDiffusion’s model architecture is divided into two modules: structure, which captures geometric aspects, and sequence, which captures intra-sequential correlations. The structure module extracts characteristics from 3D RNA backbone structures via geometric deep learning approaches, while the sequence module is used to model sequence relationships by applying transformer blocks. Both conditional and unconditional sequence distributions are modeled during training to increase versatility. Sequence sampling is accomplished via a generative denoising technique based on reverse-time stochastic differential equations (SDEs) parameterized by the data prediction model. Sequences are generated by combining ancestral sampling with self-conditioning. Parameters such as noise schedule and distribution weighting affect the model’s performance in balancing sequence recovery and variety.
Results Performance Metrics
The scientific evaluation of RiboDiffusion, which is dedicated to implementing RNA inverse folding for secondary structure, is impeccable. This is an assortment of 3D RNA structures from the PDB database and RhoFold-predicted 3D structures included in the library. Seeing the structural features for clustering purposes, the researchers separate structures into training, validation, and test sets depending on their sequence and structure similarities. RiboDiffusion is tried in competition with four machine learning baselines and secondary structure-based aptitude and gets top marks in terms of recovery rate among various clustering setups. Its advantage is obvious when it is stronger than baselines, especially in such cases with RNA in which there are more sequence or structural differences from training samples. The additional probe has disclosed that it is equally effective in all RNA lengths coverage, with the efficiency declining to a certain extent for the shortest RNA type. Finally, the model performs cross-family analysis in combination with in-silico folding calibration.
Concluding Remarks
The RiboDiffusion model plays an integral role in disclosing the puzzle of RNA folding and thus stands as the embodiment of development in molecular biology. The research findings concerning RNA’s structure not only enlighten humans but also broaden the horizons of molecular design and synthetic biology. The RiboDiffusion model is proof that informational models bring us closer to the understanding of mysteries of the molecular world even though we can’t observe them directly by the naked eye, giving us the impetus to explore, observe, and innovate.
Article Source: Reference Paper | The source code is provided at GitHub.
Important Note: bioRiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.