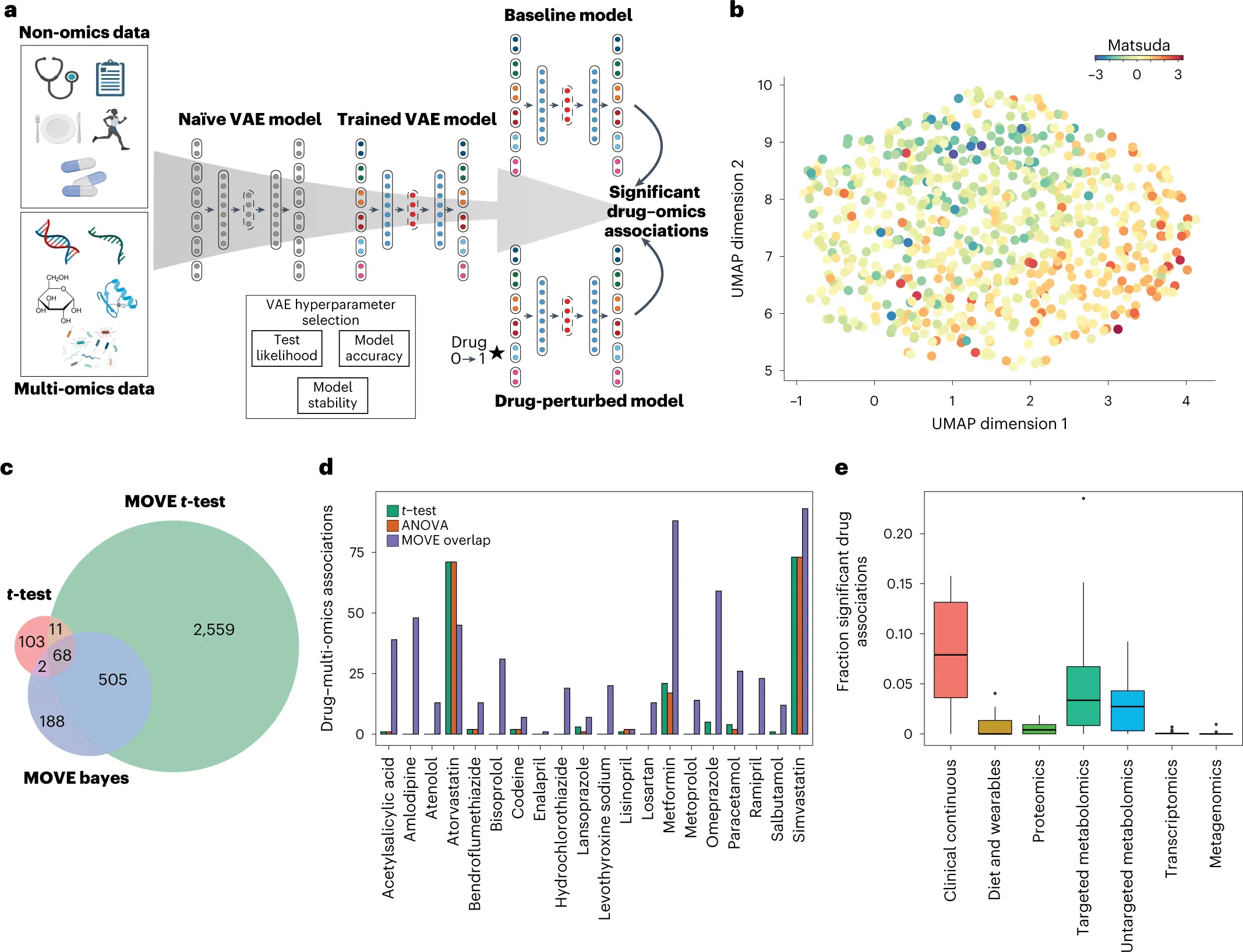
Multiple omics technologies may be used in biological cohorts to identify patient-level disease features and unique treatment responses. The size and heterogeneity of the multi-modal data, however, constitute integrating and inference a challenging challenge. Multi-omics variational autoencoders (MOVE), a deep-learning-based framework created by researchers from the University of Copenhagen and Copenhagen Research Center, was used to integrate such data on a cohort of 789 individuals with newly diagnosed type 2 diabetes who had deep multi-omics phenotyping from the DIRECT consortium.
Drug-omics connections for the 20 most common medications prescribed to persons with type 2 diabetes have been found across the multi-modal datasets using in silico perturbations, with much higher sensitivity than univariate statistical analyses. These allowed for the discovery of brand-new connections among metformin and the gut microbiota as well as the two statins’ opposing chemical reactions, simvastatin and atorvastatin. Drug effects are dispersed over the multi-omics modalities, according to relationships to measure drug-drug similarities and evaluate the degree of polypharmacy.
People with complicated disorders like type 2 diabetes (T2D) exhibit diverse drug-response patterns. Comorbidities and polypharmacy are typical contributory factors, along with many organs and confounders. On the other hand, a person’s molecular profile might vary significantly as a result of receiving therapy with one or more medicines and the related polypharmacy consequences; however, these alterations are still completely unexplored. Deep phenotyping and multi-omics screening are becoming more widely available, which has been helpful in characterizing T2D and other diseases and provides the chance to learn more about how medications work to treat disease processes.
For examining links between medications and molecular phenotypes, cohort studies may be very helpful. They can also be used to modify the design of randomized control trials to examine direct causal correlations. Univariate statistical techniques, linear and logistic regression, dimensionality reduction, and clustering analyses are frequently used in cohort data analysis techniques. However, when applying similar studies to multi-omics data, they become more complex, and conventional approaches to data interpretation fall short of using the full potential of multi-modality data.
Examples of methods that may integrate many modalities are multi-omics factor analysis (MOFA), iCluster, and data integration analysis for biomarker identification utilizing latent components (DIABLO), all of which are implemented in mixOmics. However, the main goal of these techniques is to identify components or latent variables that may be utilized for illness visualization, grouping, or prediction.
In this article, integration and extraction from a cohort of T2D patients who have been thoroughly phenotyped using unsupervised deep learning are demonstrated. The developers of this deep-learning model went beyond this by exploiting the generative capability of VAE models, whereas previous approaches for vertical integration of multi-omics data focus on encoding the data to components or latent representations that can be utilized for grouping and classification. MOVE may find significant drug-omics connections for a larger range of medications than conventional univariate statistical analyses. Findings show that these advancements result from the generative models’ increased capability as a result of their capacity to infer multi-omics alterations for people not taking medicine.
Identifying molecular connections and treatment results
It is challenging to determine the directionality of medication connections, and the causal inference process is further complicated by the cross-sectional design and clinical data-driven medical judgments. As a result, it is impossible to determine if a drug’s effects are caused by it; nonetheless, the data can be used to create informed studies and randomized clinical control trials. By examining the long-term impacts and connections, further extension using longitudinal multi-omics data and modeling time might provide additional knowledge about the causation of the medications.
Similarly, this technique allows for personalized patient analysis in an N-of-1 approach. It is well known in the medical field that choosing a medicine or therapy at the same time typically precludes carrying out the control experiment of utilizing a different medication. Although the cohort size is small, this method may be effective in identifying molecular connections and treatment results for specific individuals in bigger cohorts of tens to hundreds of thousands of patients.
Outcomes of the study
Creating a VAE for the integration of multi-omics data using a dataset of 789 T2D patients who had just received a new diagnosis and had rigorous multi-omics characterization. With the exception of metagenomics data, where two-thirds of the people (532) lacked any data, the data had an overall average of 8,807 variables per individual, and the median missingness within each omics dataset was less than 5%.
Important clinical indicators are present in the latent space and examining the neural network weights linked to the input variables of the encoder show how well the model represented the structure of the clinical data.
Drug extraction for clinical and omics correlations focused on whether the model had picked up on relationships between the clinical, pharmaceutical, and multi-omics data. A method based on randomly perturbing input characteristics was created. For instance, mimicking the drug to each of the people who did not get it in order to find connections between a certain drug and all other qualities. The same therapeutic medication class in the anatomical therapeutic chemical categorization (ATC) system was also eliminated, in addition to people who were already receiving the treatment.
Findings of drug and multi-omics correlations via MOVE
The MOVE methodology was then used to uncover drug connections in the DIRECT multi-modal data. The 3,143 and 763 significant connections to the multi-omics and clinical traits, respectively, were found using the two approaches, MOVE t-test and MOVE Bayes.
Metformin was linked to changes in T2D biomarkers
Metformin is substantially correlated with 12 clinical T2D indicators, including blood pressure, insulin clearance, active GLP-1, glucose levels from the mixed-meal glucose tolerance test, and glucose sensitivity.
Metformin and omeprazole’s interaction with the gut microbiome
Recent research has demonstrated how drug use can alter the makeup of the human gut microbiota. This led to the discovery of two medications—metformin and omeprazole—that had meaningful relationships with the metagenomics data, increasing the number of eleven metagenomics species while decreasing the number of six other species.
The two statins, simvastatin, and atorvastatin, which are often used to treat high blood cholesterol by reducing low-density lipoprotein, were linked to lower levels of cholesterol and low-density lipoprotein.
Concluding interpretations
Finally, this study aims at emphasizing that this approach is not limited to drug linkages; in theory, all omics data may be investigated for links across datasets. As a result, it is believed that the generative method opens up new possibilities for the analysis of massive amounts of multi-omics data in order to find potential new biomarkers and look into potential direct drug effects in highly dimensional molecular data that lead to testable hypotheses. The two statins had various effects on the omics data when examining the multi-omics relationships. As a result, correlations between the medications and multi-omics data were discovered, with metformin being one of them, highlighting the significance of vertical integration.
Article Source: Paper Reference
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Riya Vishwakarma is a consulting content writing intern at CBIRT. Currently, she's pursuing a Master's in Biotechnology from Govt. VYT PG Autonomous College, Chhattisgarh. With a steep inclination towards research, she is techno-savvy with a sound interest in content writing and digital handling. She has dedicated three years as a writer and gained experience in literary writing as well as counting many such years ahead.