Despite the immense hurdles posed by cancer, technological developments, particularly in medical imaging and biomarker identification, provide a ray of hope. Foundation models have revolutionized cancer imaging biomarkers, delivering unprecedented accuracy and insights vital to early identification, evaluation, and therapy. This research used a deep convolutional encoder as its foundation model, first pre-trained by comparing volumes with and without lesions. The model’s clinical application entailed collecting biomarkers and analyzing them across three classification tasks using varied datasets. The implementation involved two approaches: training a linear classifier on extracted features and fine-tuning the model parameters using transfer learning. The foundation model was evaluated against supervised models that were initialized randomly and those that used transfer learning, and it was benchmarked against openly available modern models like Med3D and Models Genesis using a rigorous approach. Several factors, including quantitative performance, stability, biological analysis, and efficiency, were carefully evaluated across a variety of application situations. The primary goal was to discover innovative biomarkers that may catalyze advances in both the scientific and clinical sectors, thus speeding up progress in understanding and treating medical problems.
Introduction
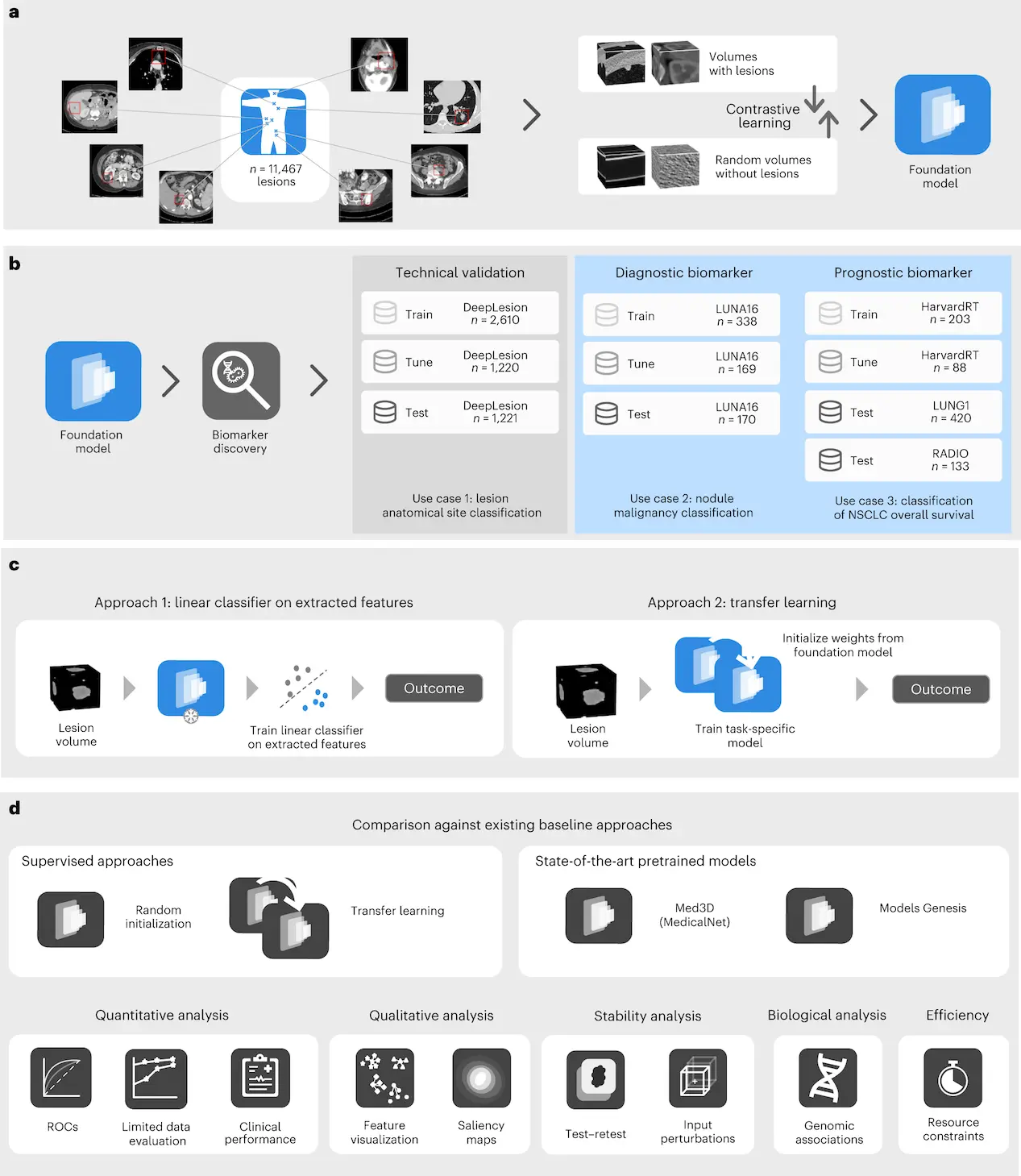
A study published recently in Nature Machine Intelligence by (Pai et al., 2024) explained a novel approach for the identification of cancer imaging biomarkers using foundational models. Biomarker detection can be built upon this foundation model for cases with limited datasets. The team of researchers used a combination of five different datasets, including DeepLesion, LUNA16, LUNG1, RADIO, and HarvardRT publicly available and internal data to train and test the foundation model whereby DeepLesion comprises multiple lesions from different patients’ cases, whereas LUNA16 and LUNG1 are used to validate diagnostic as well as prognostic imaging biomarkers. Permission to access the HarvardRT dataset containing confidential protected health information is required through formal channels. They carried out an extensive analysis and comparisons with baseline approaches so as to demonstrate how their strategy contributes towards improving the accuracy and reliability of cancer imaging biomarkers. The study developed a foundational cancer imaging biomarker identification model based on 11,467 radiographic lesions. It proved more effective than prior techniques, especially when limited data were involved due to its stability and strong connections within biological systems. Results showed efficiency as well as possibilities for broad clinical usage.
Understanding cancer imaging biomarkers
Biomarkers for cancer imaging are measurable indicators of biological processes found using techniques such as MRI, CT, and PET scans. They give fundamental data about cancer presence, extent, aggressiveness, and treatment response. These biomarkers encompass tumor size, shape, texture, vascularity, and metabolic activity. Previously manually analyzed, AI and machine learning have transformed the process to faster, unbiased analysis with less subjectivity and errors.
Methodology used in this study
Data Collection: This involved five different datasets, including Deep Lesion, which covers various lesion types that have been annotated over two decades.
Model-Implementation: A basic model was developed and then compared against previous methods for the categorization of lesions’ anatomical sites based on balanced accuracy and mean average precision.
Survival-Analysis: Kaplan Meier curves, hazard ratios, and univariate Cox regression were employed in LUNG1 and RADIO datasets to study survival for NSCLC prognosis.
Feature visualization: For high-dimensional feature analysis, t-stochastic neighborhood embeddings and principal component analysis were combined with saliency maps per task generated by fine-tuned base models.
Model Stability Tests: Model stability was assessed through test-retest and inter-reader variance evaluations to affirm the trustworthiness of the results.
Results
- About Lesion Anatomical Site Classification (Case 1)
The fine-tuned Foundation model had an average mAP of 0.857 and a balanced accuracy of 0.804, which is way better than the baseline methods. In addition to this, features from the foundation model outperformed other baseline techniques in terms of equal precision and mAP, even under simple linear classification. The foundation model extracted meaningful data for lesion anatomical location categorization as indicated by well-defined clusters and interpretable features on visual inspection.
- Nodule Malignancy Prediction Results (Case 2)
This study investigated the prediction ability of malignancy using 507 lung nodules from the LUNA16 dataset. Different models were trained on another test set of 170 nodules to evaluate generalizability. AUCs and mAPs were used to gauge nodule malignancy prediction performance, and different models performed exceptionally well.
- On NSCLC Prognostication (Case 3)
The present study utilized Kaplan-Meier curves to conduct NSCLC prognostication survival analysis based on the LUNG1 and RADIO datasets. Univariate Cox regression assessed the hazard ratios for each intervention plan, and both datasets showed significant differences between groups.
Stability of the model
The reliability of the foundational model was appraised through test-retest scenarios and inter-reader variability assessments. Intriguingly, there was great steadiness in the model with intraclass correlation coefficient (ICC) values of 0.984 and 0.966 across a number of iterations. Moreover, this showed very good stability against simulated inter-reader variances in feature differences as well as prediction accuracy.
Discussion
This work has shown that a foundation model trained by self-supervised contrastive learning can predict anatomical location, malignancy, and prognosis in many cohorts very effectively. Importantly, this model performed well in anatomical lesion location categorization as well as malignancy prediction, especially under low data availability circumstances. Furthermore, cancer-related applications benefited from accurate imaging biomarkers derived from features generated from the foundation model, even with smaller training datasets.
Implications and Future Directions
Findings from this study have important implications for cancer research and clinical practice. The researchers want to encourage more investigation and cooperation in the field of cancer imaging biomarkers by publicly disclosing their foundation model and replicable methodology. This inclusive strategy has the potential to unearth unique insights that may dramatically enhance patient care and treatment results. Through collaborative initiatives, the profession can benefit from varied viewpoints and collective experience, perhaps leading to breakthroughs in understanding and resolving cancer-related difficulties.
Conclusion
Foundation models represent a paradigm change in cancer imaging biomarkers, offering unparalleled capabilities for early detection, diagnosis, and therapy planning. We are obtaining new insights into cancer intricacies by leveraging artificial intelligence and machine learning, which is revolutionizing detection and management techniques. With continued innovation, the horizon seems optimistic for patients and physicians, as foundation models pave the way for a future in which cancer is prevented and curable rather than simply treated.
Article Source: Reference Paper | Pipeline used in this study can be accessed either from the AIM webpage or directly from GitHub.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.