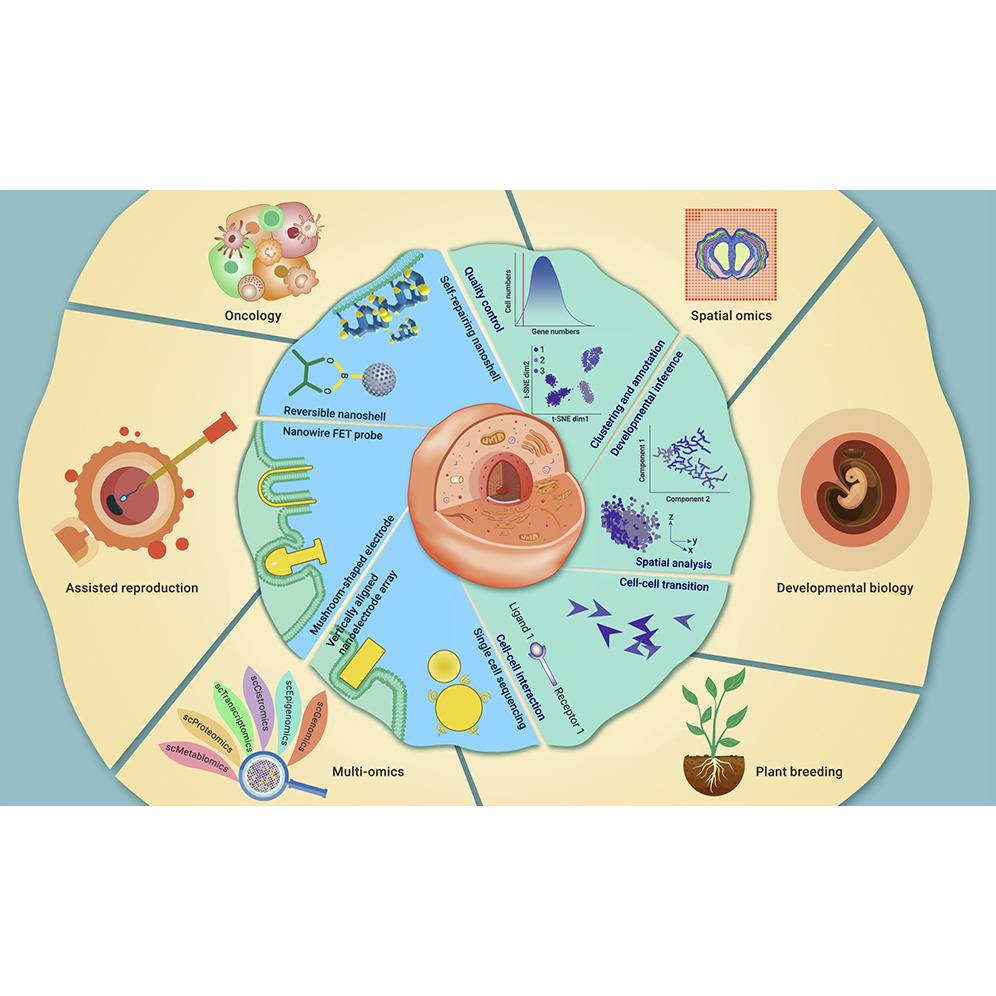
Researchers from China examined the experimental and bioinformatics approaches used in single-cell research as well as their applications in a variety of industries, thereby predicting future developments in single-cell technology.
A growing number of single-cell technologies have been introduced in past years. Large-scale initiatives like the Human Cell Atlas, Mouse Cell Atlas, Mouse RNA Atlas, Mouse ATAC Atlas, and Plant Cell Atlas have fabricated an enormous quantity of single-cell omics data. Numerous bioinformatics techniques for quality standards, cluster analysis, cell-type categorization, evolutionary hypothesis, cell-cell transition, cell-cell interaction, and spatial analysis have been created based on this single-cell big data. The molecular environment at the single-cell level can be uncovered using cutting-edge experimental single-cell technologies and large data analysis techniques based on artificial intelligence.
In addition to reviewing the experimental and bioinformatics approaches used in single-cell research, the aspects of employing diverse contexts and futuristic goals and projects of single-cell technologies are also needed to be encompassed. The Human Genome Project’s 2003 completion marked the commencement of the biological interpretation of the three billion nucleotides that make up the human genome.
The single-cell RNA sequencing (scRNA-seq) experiment was first reported by Tang et al. in 2009, which marked the beginning of the single-cell omics sequencing field of study. Over the years, method development has accelerated. Numerous scRNA-seq tests based on various RNA capture and cDNA amplification techniques have been published. The Linnarsson group discovered the high multiplex power of scRNA-seq in 2011, and the microfluidics droplet and microwell technologies further improved it in 2015. Even more, cells could be processed at a throughput of hundreds or tens of thousands of cells using the in situ multimodal semantic web technique.
Numerous omics, including the genome, epigenome, transcriptome, and others, are present in a single cell. In 2013, single-cell HiC and single-cell DNA methylome sequencing tests, two single-cell epigenome sequencing methods, were developed. The Greenleaf group created the scATAC-seq (single-cell sequencing assay of transposase accessible chromatin), which was promptly converted to single-cell analysis in 2015. It is now the fastest-growing method.
Multiple omics may be measured simultaneously in a single cell using single-cell multi-omics sequencing methods. Assays to concurrently detect the genome and transcriptome were published in 2015, while those to simultaneously detect the DNA methylome and the transcriptome (together with genome CNVs) were published in 2016. Recently, numerous high-throughput techniques for concurrently measuring transcriptome and chromatin accessibility were published.
There have been three documented sets of spatially resolved transcriptomics techniques.
- The Cai Long group identified sequential barcoding single-molecule RNA fluorescence in situ hybridization (seqFISH) in 2014, and the Zhuang Xiaowei group improved it in 2015. (MERFISH).
- The Nilsson group utilizing padlock probes reported the second set, in situ sequencing, in 2013.
- The Lundeberg group provided a description of the third set, known as in situ capturing, in 2016.
Since their inception, these strategies have been evolving, which will be covered in more depth later in this study.
Recent years have witnessed immense development in the arena of revealing a good number of Single-cell technologies
These include;
- Single-cell surface functionalization,
- Intracellular electrophysiological,
- Single-cell isolation.
- Single-cell sequencing and
Despite being the most popular and commonly utilized single-cell technology, single-cell isolation is necessary for the development of single-cell sequencing.
Single-cell surface functionalization, in which a single cell is coated with nanostructured materials, has been created to improve the viability and function of the cell and equip the cell with abiotic activities that are entirely different from its original specializations. This technique was inspired by the properties of cells in living organisms. Multiple apical dendrites in a single neuron and stable recording and interrogation of the same individual neurons in neural networks hold the promise of aiding in the understanding of complex neurological disorders that progress over time, advancing the complex brain functionality with higher resolution, and potentially enabling “cyborgs” into the real world. The integrity of automation has boosted the quantitative evaluation of cells to several thousand High-throughput cell sorting has recently been made possible by the development of nano-droplet and pico-well technologies, which can separate tens of thousands of cells at once. Many industrialized high-throughput single-cell sequencing systems, such as New VASA-seq and SORT-seq, have been created based on these single-cell separation methods.
Assessment of single-cell sequencing data via bioinformatics
ScRNA-seq data were the most developed and widely used high-throughput single-cell data. The conversion of scRNA-seq data into biological and medical information and subsequent uses depends heavily on algorithm and software development in bioinformatics. Due to the vast diversity of scRNA-seq applications, it is important to keep in mind that none of the bioinformatics tools now in use are universally applicable. So, in this section, we give a quick overview of the ideas that are important for choosing the right bioinformatics tool for this study.
- Single-cell sequencing can illustrate cellular and molecular features.
- The arrangement of cells and their correlations may be reconstructed using spatial omics.
- Single-cell multi-omics will provide a complete picture of the cell.
Monitoring and evaluation
One cell’s mRNAs cannot be completely collected and sequenced due to the low RNA concentration of single cells and the stochastic nature of existing scRNA-seq methods. Three often used criteria for traceability during scRNA-seq data processing include the upper and upper gene counts, as well as the proportion of mitochondrial genes. Controlling the frequencies of empty cells, doublets or multiplets, and cells in poor states is the goal of quality control. It is important to remember that these quality control criteria vary amongst biological specimens, hence there are no uniform cutoffs.
Ongoing research based on single-cell technologies
The expansion and implementation of scRNA-seq in the post-genome era have added a cellular dimension to omics data in addition to the molecular one. The Human Cell Atlas ([HCA]) has been started to build cellular reference maps for the location, function, and properties of every cell type in the human body, much as the Human Genome Project (HGP), which helped biomedical science make the transition to an omics age. A new step forward in biomedical innovation will result from the successful completion of a human reference cell map across the body. The National Cancer Institute-funded Human Tumor Atlas Network and the National Institutes of Health Common Fund’s Cellular Senescence Network are two more focused single-cell initiatives of cellular senescence and tumor.
Incorporating single-cell technologies in prominent research areas
Although there are other single-cell technologies, single-cell sequencing is the most often utilized. Oncology, immunology, plant breeding, and reproductive and developmental research are the main fields in which it is used.
- Using single cells to sequence tumors and immune systems
- Single-cell sequencing in IVF technology
- From the embryonic through the fetal phases of human development, gene expression patterns are bridged by single-cell data resources.
- Plant breeding via single-cell sequencing
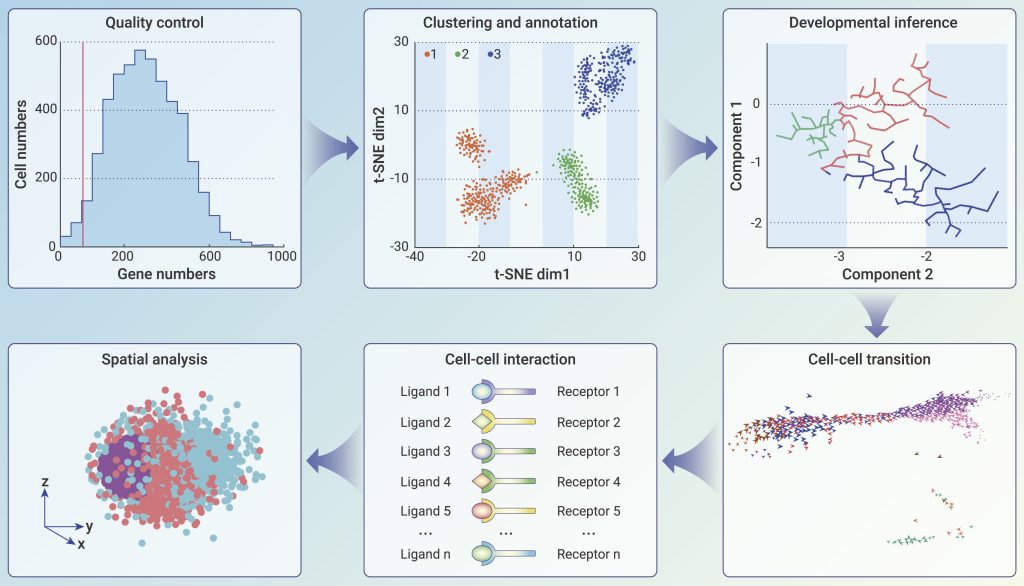
Image address: https://doi.org/10.1016/j.xinn.2022.100342
Final thoughts on Single-cell technologies
Single-cell technologies shed light on the expectation that they will serve as the foundation for many investigations. In the age of the single cell, new cell kinds and molecular mechanisms will be discovered, much to how new species and their evolutionary paths were discovered during Darwin’s time.
Article Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Riya Vishwakarma is a consulting content writing intern at CBIRT. Currently, she's pursuing a Master's in Biotechnology from Govt. VYT PG Autonomous College, Chhattisgarh. With a steep inclination towards research, she is techno-savvy with a sound interest in content writing and digital handling. She has dedicated three years as a writer and gained experience in literary writing as well as counting many such years ahead.