A team of researchers at the Eric and Wendy Schmidt Centre at the Broad Institute of MIT and Harvard developed a model that uses kernel methods to facilitate transfer learning. Kernel methods are simple machine learning algorithms that are used to train models on data that has been transformed beforehand. Transfer learning is a technique in which a model is trained on a source task, and the ‘knowledge’ gained from this is transferred to a target task. In this study, the model is applied to carry out virtual drug screening, where it helps in predicting drugs that are compatible with their corresponding cell lines when limited data is provided.
Transfer Learning and Kernel Methods: An Introduction
Transfer learning utilizes knowledge obtained from a ‘source’ to bring about improvements in the target task. It has applications in the fields of biomedical sciences and computer vision. It relies on complex deep neural networks. Kernel methods are simple machine learning methods that are comparable to neural networks. After transforming the data provided to it beforehand, it performs linear regression, which contributes to the simplicity of its operation. There have been recent developments in neural tangent networks (NTKs), a combination of kernel methods and neural networks.
This model attempts to be more scalable, with one of the key objectives being to facilitate the identification of drugs without training new models for each type of cancer cell line in question. Conventional transfer learning methods require a specified source and target. By applying kernel methods, general sources can be used for targets that have variable label dimensions.
Limitations of prior approaches
One major issue with developing machine learning models that expand over various types of source and target tasks is the lack of scalability; existing models are restricted to specific tasks only. The standard mode of operation for transfer learning has been to replace and re-train the last layer of a pre-trained network in a set of neural networks. In contrast, there is no such standard operation when using transfer learning in kernels. Previous approaches to implementing this involved incorporating the same label sets for the source and the target, where it was assumed that data for both tasks was available. This increased the computational cost as the kernels were valued using matrices, and it also proved to be computationally restrictive due to the large amount of storage that was required by the dataset.
Working of the model
The kernel is trained on source data, and transfer learning is applied by carrying out two key operations:
- Projection: It involves applying a pre-trained source kernel to the dataset of the target task to make predictions. A secondary model is trained on the predictions to resolve a target task by correcting prediction errors on the second task and classifying it into the right category. This operation is most effective when the source predictions have sufficient information on the target task.
- Translation: If the same label sets are available for the source and target tasks, a trained correction term is added to the source models to fine-tune them and help them adapt to the target task. This operation is useful for virtual drug screening. Predictions of effective drugs for one cell line can be extended to predict the effect of the same drugs on another cell line. It can also be performed when information about either the source or the target task is not accessible.
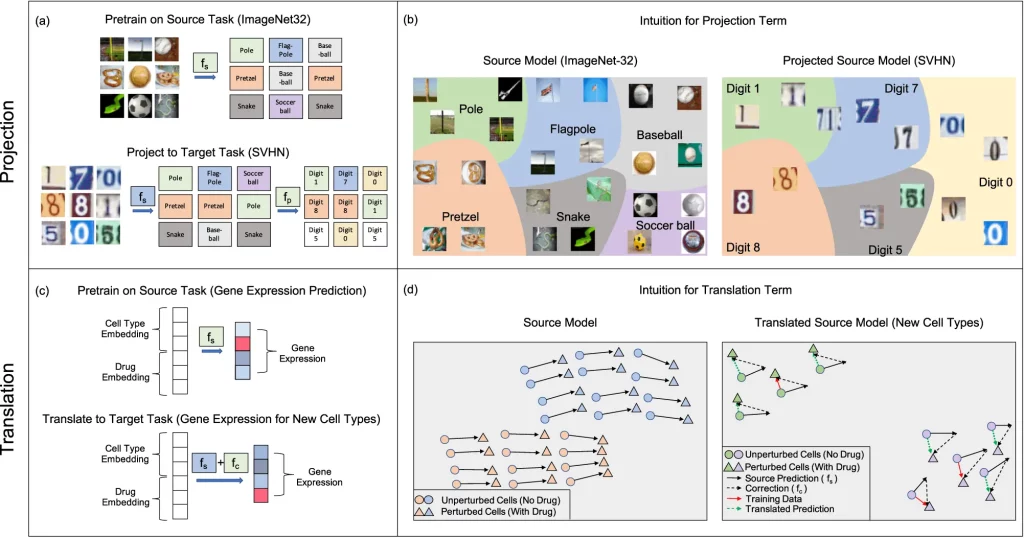
Image Source: https://doi.org/10.1038/s41467-023-41215-8
These operations are generally carried out in any transfer learning process; the only difference here is that the kernels used are flexible, simple, and more effective than neural networks. This model is also compatible with kernel regression solvers like EigenPro, which are capable of scaling large datasets. The accuracy of image classifications was increased by 10% using transfer learning methods on kernels.
It is also ascertained that the number of target samples affects the performance while modeling a target. Scalability is considered the mathematical basis for carrying out the operations as mentioned above and is determined through scaling laws. Prior algorithms used to analyze meta-learning and multitask learning algorithms did not lay out any form or method of calculating the risks associated with the predictions made by the algorithms. Such risk calculations are essential to derive scaling laws, making prior algorithms prone to becoming liabilities. In this model, risk is calculated using non-asymptotic formulas. Asymptotic methods mathematically tend to a certain limit.
Applications in Virtual Drug Screening
It is expensive and time-consuming to measure all possible combinations of cell lines and drugs experimentally. The goal here is to identify drug candidates and combinations that are worth performing experimental validation to create a more productive and efficient pipeline for drug discovery.
The key objective of applying transfer learning methods using kernels is to identify drugs without training new models for each type of cancer cell line with which it has a potential combination, saving a lot of time and resources. Two datasets were used with separate objectives for each:
- Connectivity Map (CMAP): It gives data on the genes expressed by different types of cells after treating them with specific drugs.
- Cancer Dependency Map (DepMap): It provides information on the number of cancer cells that have survived when subjected to specific drugs.
The researchers tested how well the drugs worked with different cell lines using NTKs and gene expression vectors from each cell line’s place in the model. This proved to be highly useful when no data on either the cell line or the drug was available.
The main task here was to determine the effect of the drugs on cell lines that have been newly discovered and have limited data available on them. Using this model helps in deciding how much more data would be required experimentally to get better results. Added advantages of using this model include increased interpretability, that is, the properties of the drug can be used as parameters for determining how effective the model would be in predicting its effect on cell lines and to estimate any uncertainties that may arise while performing predictions.
Conclusion
Transfer learning using kernel methods expands the scope for its application in fields like biomedical research to decide which research areas can be pursued further. This model also accounts for a distribution shift, which is useful in domains like genomics and healthcare, where shifts in cell lines and population data are a regular occurrence. It reduces memory and runtime costs as well. In the pharmacological industry, scaling laws can be useful for understanding the number of samples that may be needed for assessing complex shifts between domains, for example, when considering an organism-based shift from a mouse model to a human model.
Article Source: Reference Paper | Reference Article
Learn More:
Swasti is a scientific writing intern at CBIRT with a passion for research and development. She is pursuing BTech in Biotechnology from Vellore Institute of Technology, Vellore. Her interests deeply lie in exploring the rapidly growing and integrated sectors of bioinformatics, cancer informatics, and computational biology, with a special emphasis on cancer biology and immunological studies. She aims to introduce and invest the readers of her articles to the exciting developments bioinformatics has to offer in biological research today.










