Researchers at the University of Lucerne utilized molecular imaging to evaluate the expression of certain regions of the human genome. The imaging techniques are part of an AI pipeline that involves human-robot cooperation using deep learning-based artificial intelligence (AI) tools. This would link discrete genes to the diseases that they could possibly be responsible for. The researchers cross-referenced RNA-seq data that had been collected from a large pool of individuals to data retrieved from what the researchers term the ‘Imageable Genome.’ They discovered imageable genes that can serve prognostic and diagnostic purposes.
Why is molecular imaging important?
The main goal of modern medicine is to understand the underlying causes of human diseases on a molecular level in order to curate the appropriate therapeutic method targeted toward specific diseases. To accomplish this, it is important to make sure that any treatment or cure that has been proven to be effective in a laboratory setting is moved forward to be tested in clinical trials and to ensure further that the required medication is delivered to those patients who need it the most as soon as possible. This work has been made easier through the use of various methods of molecular imaging.
Molecular medicine made its inception with a seminal paper on sickle cell anemia presented by Linus Pauling and has since accelerated the advent of molecular medicine in treating human diseases as well as preventing them. We learned more about the genetic causes of diseases when the Human Genome Project (HGP) finished decoding the entire human genome in 2003.
Molecular imaging is a method that can effectively gauge the expression of target molecules on subcellular and cellular levels in a reproducible,non-invasive, and quantitative manner. It plays a crucial role in the development of medicinal treatments to cure many diseases that affect humans.
Positron emission tomography (PET) is one such method of molecular imaging, which makes use of methods like magnetic resonance imaging (MRI) to spot molecular targets in a clinical setting, and it is a method of choice for most medical professionals. It is widely used in medical domains like oncology and neurology due to its ability to conduct serial exams of the whole body with no sampling errors. Oncology has greatly benefitted from the combination of PET, computer tomography (CT), and the use of radiotracer fluorine-18-fluorodeoxyglucose (FDG); collectively, this is known as PET-CT, and it is extensively applied in clinical diagnostics.
The relevance of radiotracers in molecular imaging
Accurately targeting key molecular markers can aid PET in uncovering its peak potential when identifying the progression of individual diseases, visualizing spatiotemporal pathobiology, and indicating transformations that generate drug resistance within genes. This targeting is currently carried out by existing radiotracers, the data for which is compiled in the National Institute of Health’s (NIH) Molecular Imaging and Contrast Agent Database (MICAD). Radiotracers can open the doors towards developing and incorporating personalized medicine as a normal method of carrying out clinical practices. Recent efforts have led to the development of radiotracers that can target specific cells present in the tumor microenvironment; recent examples of such cells are the fibroblast activation protein inhibitor (FAPI) and the prostate-specific membrane antigen (PSMA).
An unfortunate reality, however, is that fewer than 1% of the aforementioned radiotracers have made it to an actual clinical setting. As useful and promising as they may be, they are barely being utilized in clinics in the real world. One possible reason for this disappointing outcome is the lack of general awareness of the potential that certain molecules can hold in treating diseases when targeted by radiotracers, with the help of molecular imaging to spot them within the genome. These molecules can play an important role in discovering antidotes for several human diseases.
To generate awareness on these topics, the researchers in this study attempted to list the molecular products derived from the genes that can be spotted using molecular imaging. They published their findings in their research article ‘The Imageable Genome’ published in the journal Nature Communications, where they described the novel pipeline curated by them, which involves deep learning as well. They also compared this data to existing genomic datasets that have been updated to verify its legitimacy. The intersection between different areas of research, such as data science, molecular imaging, systems biology, and genomics, was highlighted by the researchers in their paper.
A peak into the Imageable Genome
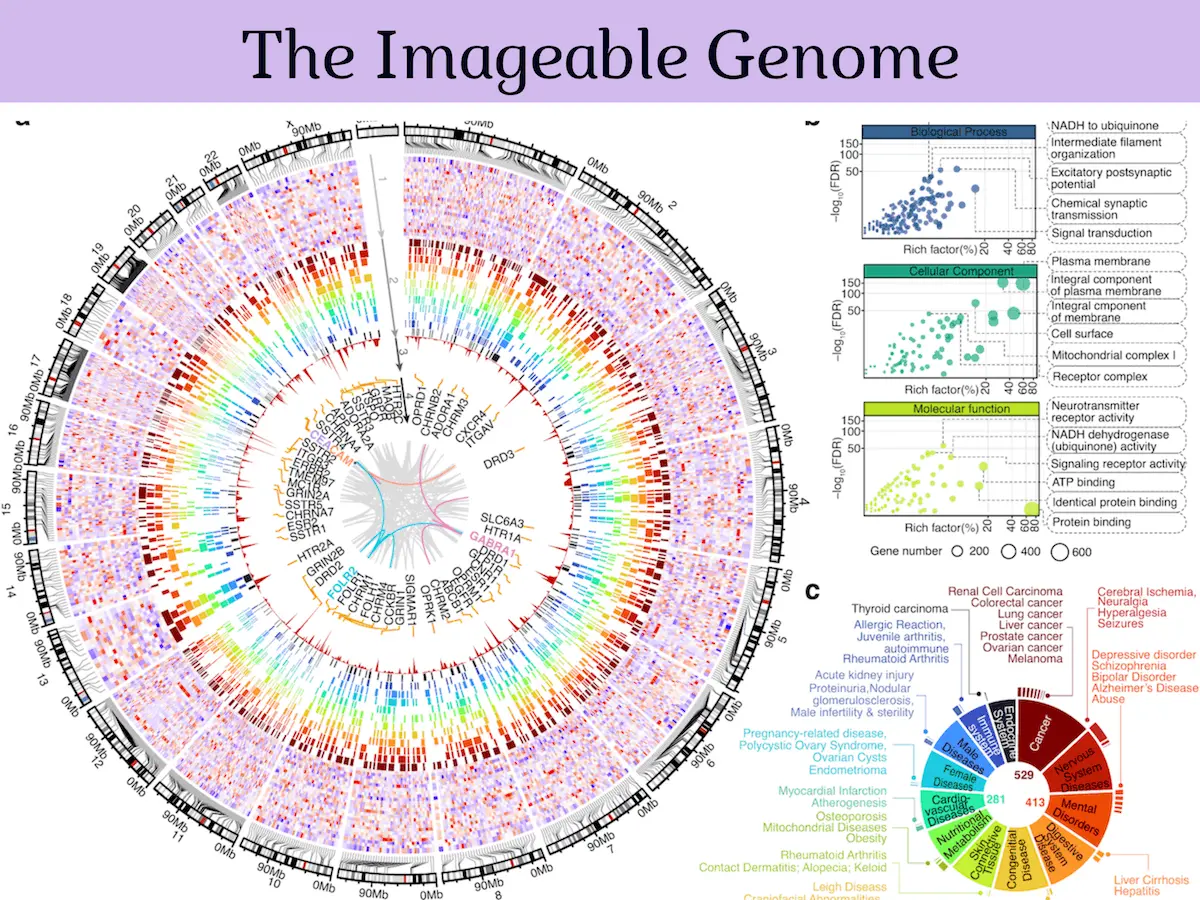
The Imageable Genome constitutes nearly 6% of the portion of the genome that codes for human proteins and 1.8% of the overall human genome, especially the regions that have their focus and critical size altered during the progression of any disease within the body. The researchers found that 2% of the genes identified by them act as markers for oncological, neurological, and cardiological diseases. They also found that the overall expression of the Imageable Genome will be affected by the development of any traits that would signal a worsening prognosis, such as the transformation of tissue from a healthy to a diseased state and any signs of resistance to treatment by drugs.
A significant challenge faced when attempting to translate molecular discoveries into clinical settings is that the discoveries based on multi-omic methods are very complex, and it is difficult to convert all that data into a language that is in line with the more simplified, decision-oriented way of dealing with data in a clinical setting. The Imageable Genome can act as a bridge to cover this gap by using molecular imaging and translating the raw data into convenient clinical imaging tests.
Conclusion
Molecular imaging is just the first break of multiple upcoming future breakthroughs in the prognosis and diagnosis of life-threatening human diseases; this is only the beginning. For years to come, researchers and medical professionals will find the Imageable Genome to be a valuable resource. The database will keep growing as long as there is ongoing research in the field of radiopharmaceuticals.
Article Source: Reference Paper | The software code Imageable Genome is available on GitHub
Learn More:
Swasti is a scientific writing intern at CBIRT with a passion for research and development. She is pursuing BTech in Biotechnology from Vellore Institute of Technology, Vellore. Her interests deeply lie in exploring the rapidly growing and integrated sectors of bioinformatics, cancer informatics, and computational biology, with a special emphasis on cancer biology and immunological studies. She aims to introduce and invest the readers of her articles to the exciting developments bioinformatics has to offer in biological research today.
















[…] Unlocking the Secrets of Disease: The Imageable Genome and the Future of Molecular Medicine […]