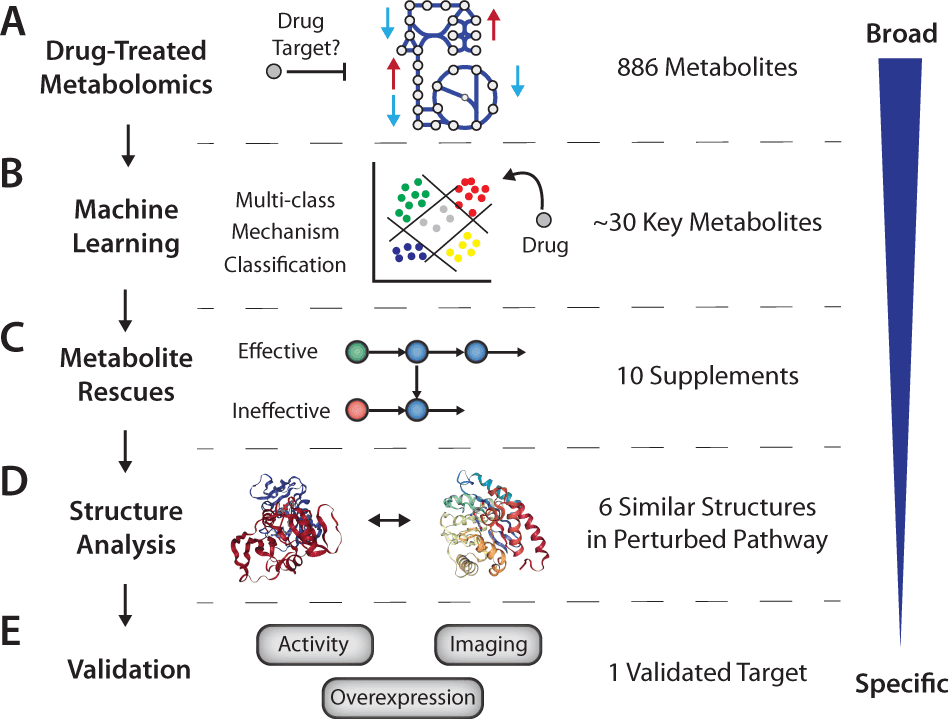
Drugs often impact intracellular off-targets, i.e., targets that are unintended. Identifying drug off-targets is extremely important for understanding the mechanism of drug action and designing effective drugs. In this context, comprehending the underlying mechanisms of antibiotics inside bacterial cells is highly required for developing antibiotics that will encounter emerging ‘drug resistance.’ A detailed workflow for investigating these aspects is critical and holds promising prospects for further studies. A recent publication from Harvard University and California University, USA, integrated omics approaches and machine learning and framed workflow to enhance drug target discovery and identify off-targets of a metabolic inhibitor.
Advancing CD15-3: Evolution-Driven Antibiotic
The researchers in their earlier study discovered CD15-3, a novel antibiotic compound that effectively targets DHFR in both wild-type and trimethoprim (TMP) resistant E. coli. However, the presence of additional interacting molecules of the antibiotic CD15-3 was suspected due to the observation that overexpression of the DHFR gene (folA) couldn’t completely recover the normal growth rate of the bacterial cell. Dihydrofolate reductase is involved in purine and thymidylate biosynthesis, important constituents of DNA. To design CD15-3 as an “evolution-drug” to tackle antibiotic resistance, comprehending non-DHFR drug targets is important. Evolution drugs can escape the evolutionary route of drug resistance in pathogens.
Insights from Metabolomic Analysis
Metabolomic analysis was performed to point out the effects of the antibiotic CD15-3 in an E. coli strain at different times of its growth. For this, the bacterial cells were treated with the antibiotic, and Mass spectrometry and Chromatographic techniques were performed, executing the standardized protocol followed by Data analysis employing several software packages.
Some significant statistics and data were inferred using machine learning models, that are: As the pyrimidine biosynthesis pathway unfolded, thymidine, an essential element, underwent a remarkable decrease in abundance, dropping by 15-fold during the mid-exponential phase and a further 17-fold in the late log phase. Precursor in the THF (Tetrahydrofolate) biosynthesis showed 12-fold upregulation in the mid-exponential phase with CD15-3 treatment and 15-fold higher abundance in the late log phase. Serine and glycine showed 20-fold lower abundances at the late log phase under CD15-3 treatment, indicating that the folate pathway is perturbed by the antibiotic.
Growth Recovery and Insights from Gene Overexpression Analysis
The researchers trained a multi-class logistic regression (LR) model to identify metabolomic perturbations associated with mechanisms of antifolate, cell membrane, DNA synthesis, translation, and oxidative stress and visualized the data with Uniform Manifold Approximation and Projection (UMAP). Clustering the projection revealed that CD15-3 shows its characteristics as an antifolate at early time points, it shows a broader growth inhibitory response. Next, key metabolites for antifolate activity were searched for with this model.
Supplements that can retrieve the growth of the antibiotic-treated cells were selected from previous analyses to understand which pathways are directly affected by CD15-3. The major observations validated by the computational analysis were supplementation with thymidine, NAG, serine, and IMP resulted in improved growth rates. Supplements that do not reduce folate deficiency displayed negligible effects on the growth rates, such as when aspartate, glycine, uridine, citrate, orotate, and AMP were added as supplements. It can be interpreted that the metabolite supplements that attenuate folate deficiency were directly linked with recovery growth when added externally.
The idea of the folate cofactor drain mechanism of CD15-3 was extrapolated by implementing constraint-based metabolic modeling, demonstrating that the alternate target of CD15-3 is also linked with the same metabolic cascade as of DHFR because metabolic modeling consistently depicted that folate inhibition is the signature perturbation induced by the antibiotic. Genome-scale Model with Protein Structures analysis showed that other enzymes don’t possess similar structures to DHFR. A comparison of active sites with the FATCAT algorithm and isomif analysis suggested possible alternative binding targets for CD15-3 in the same metabolic pathways are MTHFC, HPPK2, and DHPS.
An overexpression experiment was executed to check which of the suspected candidate could rescue growth and thus interacts with the antibiotic. Among the suspected molecules, overexpression of the folK gene that encodes HPPK2 can consistently recover growth in a broad range of contrived conditions, elucidating that HPPK also can bind with the antibiotic CD15-3. Moreover, the observation corresponds to rigid molecular-docking analysis. Where CD15-3 treated strain displayed cellular filamentation, cells with overexpressed folK gene didn’t display any morphological alterations. Further, in-vitro assays were performed to support this interpretation that CD15-3 inhibits HPPK.
An Innovative Workflow for Drug Off-Target Discovery
The study illustrates how machine learning approaches can be complemented with drug mechanism analysis to discover off-targets. Incorporating constraint-based metabolic modeling with machine learning in exploring large-scale metabolomics data assisted in apprehending metabolic perturbation signatures and narrowing down supposed pathways of drug off-targets.
The proposed framework of such studies, without even an initial idea of the drug target, can be:
- Metabolomics with comparison to previously characterized antibiotic compounds
- Supplementation experiments to explore growth-inhibited pathways
- Overexpression of candidate targets to monitor growth rescue.
Conclusion
Now CD15-3 can be deciphered as a multivalent drug, inhibiting two molecular targets (DHFR and HPPK) inside bacterial cells. Therefore, the antibiotic can prevent the phenomenon of multi-drug resistance by promoting more antibiotic stress inside bacteria as it can target two molecules essential for bacterial survival and growth. Although the framework was designed on antimetabolite targets of antibiotics, the scientists mentioned its prospects in developing a workflow for other types of antibiotics. Another crucial facet is that the research points out the foundation that can be implemented in further valuable studies, such as investigating off-targets of cancer drugs that lead to toxicity.
Article Source: Reference Paper
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.