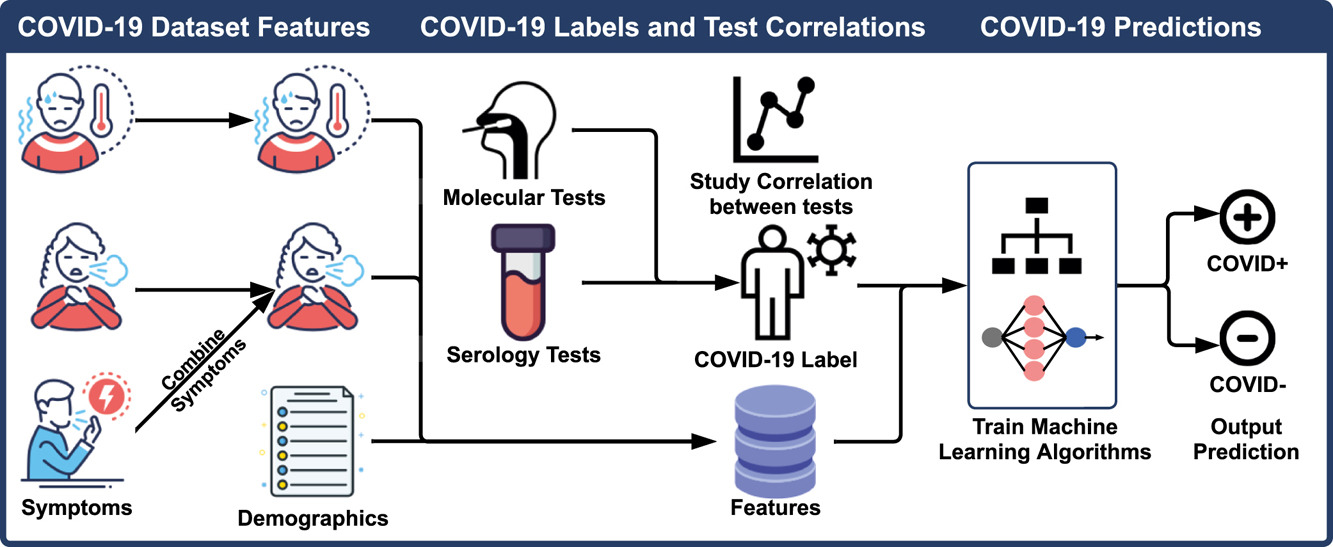
A study from Florida Atlantic University used machine learning to provide new evidence for understanding how molecular tests and serology tests are correlated and what features are most useful in COVID-19 testing. This could be useful in helping healthcare providers prioritize testing and treatment for patients most likely to have the virus.
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)-related new coronavirus illness (COVID-19) first appeared in Wuhan City, China, in 2019, and it spread swiftly around the world. Over 515 million COVID-19 cases and more than 6 million fatalities have been reported worldwide as of May 2022. The major way that COVID-19 infection spreads from an infected person to a non-infected person is by aerosol droplets from their coughing or sneezing. Asymptomatic people may potentially transmit the disease. SARS-CoV-2 has an incubation period of 2 to 14 days, with most people showing symptoms within 12 days after infection.
It is advised to get confirmatory COVID-19 testing done as soon as symptoms appear. It needs a massive diagnostic testing effort to control COVID-19 outbreaks. It has been demonstrated that testing, quarantining positive people, tracking contacts, and vaccinations are effective ways to stem the spread of COVID-19.
The symptomology of COVID-19 ranges from minimal symptoms to Acute Respiratory Distress Syndrome (ARDS) and can also be deadly. Common COVID-19 symptoms include exhaustion, coughing, fever, and breathing problems. Other symptoms include diarrhea, myalgia, a sore throat, congestion, or a runny nose. In COVID-19 individuals, neurological symptoms such as headaches, vertigo, reduced awareness, and/or taste and/or smell abnormalities are also often described. However, some signs can also appear in other viral illnesses and respiratory conditions, such as MERS, SARS, and influenza.
The two most popular techniques for quick COVID-19 infection testing are serology and molecular assays. While serology tests check for antibodies binding to specific SARS-CoV-2 proteins, molecular testing evaluates the presence of viral SARS-CoV-2 RNA.
Detection of Covid-19 using Molecular assays and Serology test
Molecular assays detect the presence of SARS-CoV-2 RNA from the virus. They can be carried out via transcription-mediated amplification (TMA), reverse transcription polymerase chain reaction (RT-PCR), or polymerase chain reaction (PCR). These assays magnify the genetic makeup of the virus to enable identification. The most used technique for identifying SARS-CoV-2 is molecular testing.
Immunoglobulin G (IgG), IgM, or IgA antibodies, which reflect an immunological response to SARS-CoV-2, are detected by serology tests such as enzyme-linked immunosorbent assays (ELISAs), chemiluminescent immunoassays (CLIA), and chemiluminescent microparticle immunoassays (CMIA).
There is presently no study on the link between serology and molecular testing and which COVID-19 symptoms are crucial for a positive test result. The study shows that machine learning models can predict COVID-19 infections when they are equipped using primary symptom and demographic variables. This study acknowledges the essential symptoms linked to COVID-19 infection and offers a method for quick screening and reasonably priced infection identification.
Heading toward the construction of an accurate prediction model
Outcomes show that the number of days a person has symptoms like fever and respiratory problems significantly impacts the COVID-19 test results. Additionally, research indicates that when subjected to post-symptom onset days of serology tests, molecular tests have substantially narrower post-symptom onset days (between three and eight days) (between five to 38 days). Since the molecular test monitors an active infection, it has the lowest positive rate.
Additionally, there are substantial differences amongst the COVID-19 tests, in part because the test methodologies deals majorly with the dynamic immune responses of donor and the amount of virus present in an infected person’s blood. A collection of various positive/negative results possibly occurs from the two distinct types of testing, even for the same donor.
Outcomes from 2,467 donors who underwent one or more COVID-19 tests served as the testbed for this study. By integrating symptoms and demographic information, the researchers developed a set of features for predictive modeling utilizing the five distinct types of machine-learning models. They investigated the association between serology and molecular testing by contrasting different test formats and their results. The 2467 donors were classified as positive or negative with the purpose of predicting test outcomes using the results of serology or molecular tests, and they generated symptom features to represent each donor for machine learning. They used easily accessible symptom features, combined with demographic information like age, sex, body temperature, and the number of days counted from the onset of the symptoms, to construct an accurate prediction model.
Categorizing related symptoms into bins and the data gathering procedure is fundamentally error-prone since COVID-19 causes a wide variety of symptoms. Without symptom reporting standards, the range of symptom features significantly expands. Binning technique to provide the scope of saving sample feature information while reducing the size of the symptom feature space.
Five machine-learning models
Because some donors may have several test results, the testbed specially qualifies the study of the correlation between serology tests and molecular testing as well as the consistency between each type of test.
The researchers employed five machine-learning models
- Random Forest,
- XGBoost,
- Logistic Regression,
- Support Vector Machine (SVM), and
- Neural Network machine learning methods.
Three performance metrics—Accuracy, F1-score, and AUC—were used to compare results.
Incorporating AI-based predictive modeling
Unresolved research-related concerns, further complicated with numerous perplexing impedes predictive modeling. The testbed developed is, in fact, innovative and amply demonstrates the relationship between various COVID-19 test kinds. The team devised a novel method for reducing noise in symptom data for clinical interpretation and predictive modeling. Such AI-based predictive modeling techniques are becoming more and more effective in tackling infectious illnesses as well as many other health challenges.
Further research has demonstrated that sensor data, in addition to symptom features, can help with the prediction of COVID-19 diagnosis since sensor data offer extra monitoring of each person’s health status. It was discovered that using sensor data and symptoms together had the best predicting performance. The sensor data is frequently taken by well-known smartwatches like Fitbit and Apple watches. These gadgets are still expensive to buy, and software programs are needed to collect sensor data in order to use the data.
Limitations of symptom prediction models
The subjective nature of symptom reporting is a drawback when employing symptoms for prediction models. Similar symptoms may be described differently by various subjects, and reporting symptoms may be biased. Additionally, there are restrictions on symptom documentation. While limiting the options for symptoms has the advantage of promoting more uniform replies, it also runs the risk of leaving out significant or genuine symptoms.
Deep neural language models have recently made advancements that may offer different ways to learn features from noisy symptoms without categorizing such elements. However, this would make the model harder to comprehend and jeopardize the decision’s credibility. For symptom prediction models, determining the incubation period, or determining the timing of the immune response, asymptomatic COVID-19 patients present an extra challenge. Other variables, such as exposure history or risk factors (healthcare professionals, etc.), may be highly helpful for predicting models for asymptomatic people.
Conclusion
In this work, researchers conducted a machine learning analysis of COVID-19 serology and molecular testing and thus suggested developing classification models for COVID-19 infection forecast using straightforward demographic and clinical variables. This investigation identifies many useful symptom markers that are significantly associated with COVID-19, including days PSO (post-symptom onset) and fever temperature. These prediction models attain over 81% AUC (Area Under the ROC, Receiver Operating Characteristic, Curve) scores and over 76% classification accuracy by utilizing developed bin characteristics in conjunction with five machine learning techniques. This study demonstrates that machine learning models that are trained using basic demographic and clinical variables can aid in the prediction of COVID-19 infections.
Article Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Riya Vishwakarma is a consulting content writing intern at CBIRT. Currently, she's pursuing a Master's in Biotechnology from Govt. VYT PG Autonomous College, Chhattisgarh. With a steep inclination towards research, she is techno-savvy with a sound interest in content writing and digital handling. She has dedicated three years as a writer and gained experience in literary writing as well as counting many such years ahead.