Fudan University researchers introduced DxFormer, a new automatic diagnostic framework in which each symptom is regarded as a token, formalizing symptom inquiry and disease diagnosis to a language generation model and a sequence classification model, respectively. The decoder encoder structure, an inverted version of Transformer, was utilized to learn the representation of symptoms by jointly optimizing the reinforced reward and cross-entropy loss. This computational approach improves on earlier reinforced learning-based methodologies. DxFormer increases symptom recall and achieves state-of-the-art diagnostic accuracy by detaching symptom queries from the diagnostic process.
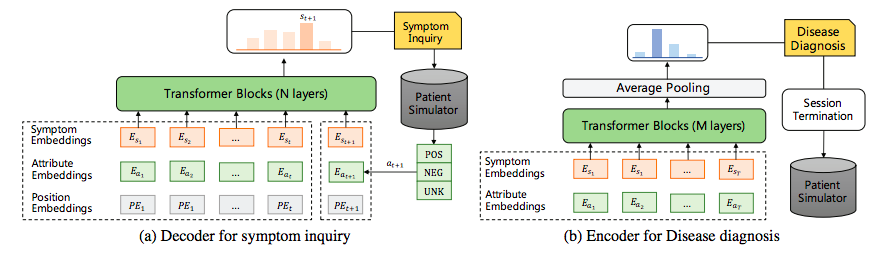
Image Source: https://doi.org/10.1093/bioinformatics/btac744
The union of the internet and healthcare provides various advantages and far-reaching positive impacts in terms of boosting service efficiency and encouraging social fairness. One of the emerging needs in this new healthcare paradigm is automated disease diagnosis, which aims to imitate clinicians’ actual diagnostic procedures. The actual disease diagnosis process can be thought of as a series of questions and responses. Doctors select pertinent questions to ask patients to have a better knowledge of the patient’s physical state. The agent in a symptom-based automatic diagnostic system can perform two types of actions: inquire about a symptom and predict an illness, that is, choose one of the elements from a fixed set of symptoms or diseases.
In recent years, there has been a surge in the study of automatic diagnosis. Most academics investigate using reinforcement learning to model this problem (RL). In RL-based systems, the AI frequently asks the patient for one or two symptoms before diagnosing. In practice, however, an average of 7 to 8 symptoms are mentioned in each talk. As a result, the agent collects insufficient symptom features, affecting illness diagnosis performance.
To address the abovementioned issues, Chen et al., present DxFormer, a decoupled autonomous diagnostic system composed of a Transformer-based decoder-encoder structure (not encoder-decoder). The decoder is used for symptom inquiry, where symptoms are handled as natural language tokens, and the symptom inquiry is characterized as a conditional text generation job. The encoder is used for illness diagnosis. The symptoms collected in symptom inquiry are fed into it as the encoder’s input sequence, with disease diagnosis modeled as a sequence classification job.
The decoder is encouraged to uncover implicit symptoms, and the encoder is encouraged to produce proper diagnoses, and the two work together independently and are trained concurrently with low interference. The experimental results show that DxFormer significantly enhances symptom recollection and diagnosis accuracy when compared to other techniques.
Significant Findings
i) MZ-4 is the first human-labeled dataset used to test an autonomous diagnostic system. MZ-4 comprises four diagnoses: children’s bronchitis, functional dyspepsia in children, infantile diarrhea infection, and upper respiratory infection.
ii) Dxy is a medical conversation dataset that has been annotated and gathered from a popular Chinese online healthcare website. Dxy contains five diagnoses: allergic rhinitis, upper respiratory infection, pneumonia, hand-foot-mouth illness in children, and pediatric diarrhea.
iii) MZ-10 is a dataset with multi-level annotations that has been enhanced from MZ-4 to include ten disorders, including common digestive, respiratory, and endocrine system ailments. MZ-10 also includes other symptoms. Medical students annotate the MZ-10. Each discussion is annotated twice, and the kappa coefficient of symptom labels is 92.71%, indicating that the two annotations are highly consistent.
DxFormer exhibited a strong performance. DxFormer can detect around 45-52% of implicit symptoms in 68 turns and achieve 64-84% diagnostic accuracy on all three real-world datasets. DxFormer significantly increases symptom recall when compared to other best baseline models, with an absolute improvement of about 12-27%. Furthermore, the diagnostic accuracy outperformed all previous models. DxFormer, for instance, boosted the accuracy by around 14 absolute percentage points above the best baseline on MZ-10.
In this paper, DxFormer, a decoupled system, employs Transformer’s decoder and encoder for symptom query and disease diagnosis with dense symptom representations. Conditional text creation and sequence classification were used to standardize symptom queries and disease diagnosis. Extensive testing using real-time datasets has shown that DxFormer can significantly increase symptom recall and diagnosis accuracy.
Limitations
Despite the positive outcomes gained, the method still has several limitations:
- In practice, agents should be allowed to question numerous symptoms simultaneously rather than a single symptom to optimize efficiency.
- Using mere symptoms for diagnosis is insufficient. More elements, such as medical examination, past medical history, surrounding environment, and so on, must be considered in real circumstances.
In the future, the authors plan to investigate automatic disease detection based on even more features—furthermore, an attempt to improve the biological interpretability of the model.
Article Source: Reference Paper | Code: DxFormer
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Shwetha is a consulting scientific content writing intern at CBIRT. She has completed her Master’s in biotechnology at the Indian Institute of Technology, Hyderabad, with nearly two years of research experience in cellular biology and cell signaling. She is passionate about science communication, cancer biology, and everything that strikes her curiosity!







