RNAs are essential components of physiological activities, however, accurately modeling their structures has long posed a challenge due to their complex structures. The National University of Singapore researchers introduced DRfold, an innovative technique to predict RNA complex 3D structures. By simultaneously learning local frame rotations and geometric constraints from experimentally solved RNA structures, DRfold addresses this issue and uses the resulting knowledge to create a hybrid energy potential that directs the construction of the structure. As RNA structural databases continue to grow, the open-source DRfold program and its effective training process offer broad applicability and prospective improvements.
RNA molecules are essential for regulating gene expression, transcription, scaffolding, and catalytic processes, among other key cellular processes. It is crucial to clarify the precise tertiary structures of RNA molecules in order to use them as pharmacological targets or to comprehend how they work within the body.
The majority of traditional RNA structure prediction techniques use simulations or homology modeling techniques. The effectiveness of homology-based approaches, such as ModeRNA and RNABuilder, for various RNA targets, is reduced since they pull structure information from previously solved homologous structural templates. Another class of techniques assembles RNA structures utilizing pieces from a prebuilt library, as shown by RNAComposer and 3dRNA. Statistical potentials are used by ab initio RNA structure prediction techniques like SimRNA and FARFAR to direct structure folding simulations. While incorporating domain expert knowledge can enhance performance, automated benchmark tests often reveal suboptimal results, highlighting the challenging nature of automated prediction for regular RNA structures.
Deep machine learning has recently shown potential in predicting the characteristics of RNA structure. Convolutional neural networks (CNNs) or recurrent neural networks (RNNs) have been used in a number of studies to improve the accuracy of secondary structure predictions for RNAs. A new tool, DRfold, a unique deep learning pipeline aiming at enhancing Ab-initio RNA structure prediction, was introduced as a result of the success of deep learning in 3D protein structure prediction. Due to the scarcity of RNA structures, DRfold uses a coarse-grained model for RNA instead of the full-atom, end-to-end training used for proteins. The phosphate P, ribose C4′, and glycosidic N atoms of the nucleobase are the only particular atoms taken into account by this model.
The Outcome Revealed
The RNA tertiary structure prediction method used in the DRfold pipeline is structured. A query sequence and its anticipated secondary structure (SS) are entered to start the process. An embedding layer uses this data to create crucial sequence and pair representations. These embedded representations are processed by 48 RNA transformer blocks, supplying energy for the following processes.
The RNA transformer building blocks have two functions. First, by permitting nucleotide-wise rotation matrices and translation vectors, and second, they aid end-to-end RNA global-frame training. This learning mechanism makes it easier to regain the crucial atomic coordinates for the RNA structure. In the meantime, a different but related collection of transformer blocks is used to predict the inter-nucleotide geometry of RNA using the pair representations produced from the transformer blocks.
The predictive outputs from the previous steps, namely the frame vectors and geometric restraints, are amalgamated into a composite potential. This potential is instrumental in guiding RNA structure reconstruction through a gradient-based optimization process. The primary objective of this optimization is to identify the RNA conformation with the lowest energy, which is subsequently selected as the output model.
To enhance the precision and reliability of the models, the coarse-grained representations of the RNA structures generated by the pipeline undergo a meticulous refinement procedure. This procedure leverages a two-step molecular dynamics-based approach, which aids in atomic-level structure reconstruction and fine-tuning.
The effectiveness and performance of DRfold were rigorously evaluated through testing on a dataset of 40 non-redundant RNA structures. These structures exhibited varying lengths, spanning from 14 to 392 nucleotides, and were carefully curated from sequence cluster centers, adhering to a stringent sequence identity cutoff of 90%.
DRfold Excels Beyond Past RNA Structure Prediction Methods
In a comparative evaluation with representative control methods, including RNAComposer, 3dRNA, SimRNA, RNA-BRiQ, and FARFAR, DRfold consistently demonstrates superior performance.
The assessment involved analyzing the root mean squared deviation (RMSD) and TM-score, crucial metrics for structural similarity assessment. DRfold significantly outperforms control methods in terms of RMSD, showcasing an average RMSD of 14.45 Å, notably lower than all control methods. Furthermore, DRfold excels in TM-score, with an average of 0.435, a remarkable 73.3% higher than the second-best method, 3dRNA. Additionally, DRfold achieves a high success rate of correct folds, with 45% of models exhibiting TM-scores greater than 0.45, surpassing the best control method’s success rate of 12.5%. DRfold exhibits a superior ability to recover base interaction networks and maintain the correct chirality of RNA helices.
End-to-End Models Enhance Geometric Constraints in RNA Structure Modeling
In the DRfold RNA structure prediction pipeline, two crucial types of potentials, FAPE, and geometry potentials, are derived from separate transformer networks, providing essential complementary information. The FAPE potential facilitates end-to-end learning, directly predicting rotation matrices and translation vectors for nucleotide frames, significantly enhancing the prediction performance. When considering only geometry potentials for structure optimization, the performance drops significantly, underscoring the importance of the end-to-end potential. The end-to-end models already outperform control methods, showcasing their efficacy. Moreover, incorporating end-to-end and geometry potentials collectively improves the quality of DRfold models, demonstrating the synergy of these complementary approaches. The results emphasize that even though they stem from the same initial data, these potentials provide unique structural insights that improve the accuracy of RNA structure predictions.
Incorporating Secondary Structure Prediction for Enhanced Feature Learning and Model Construction
In DRfold, utilizing predicted secondary structure (SS) is vital for accurate RNA tertiary structure prediction. Comparisons highlight a substantial drop in TM-score when SS predictions are excluded. Even sequence-free DRfold modeling, using only predicted SS, yields higher TM-scores than the best third-party method, emphasizing the predictive power of SS. Incorporating sequence-specific base-pairing significantly enhances structure quality.
Structure Refinement for Enhanced Physical Realism in DRfold Models
DRfold employs a two-step refinement process from coarse-grained to atomic-level models to enhance the accuracy and biological relevance of predicted RNA structures. The initial coarse-grained models exhibit high clash and MolProbity scores, indicating structural inaccuracies. After refinement using molecular dynamics and minimization, clash and MolProbity scores significantly improved, aligning the model quality more closely with experimental structures. The global model quality, assessed by the TM-score, experiences only a minor decrease, showcasing the effectiveness of the refinement process.
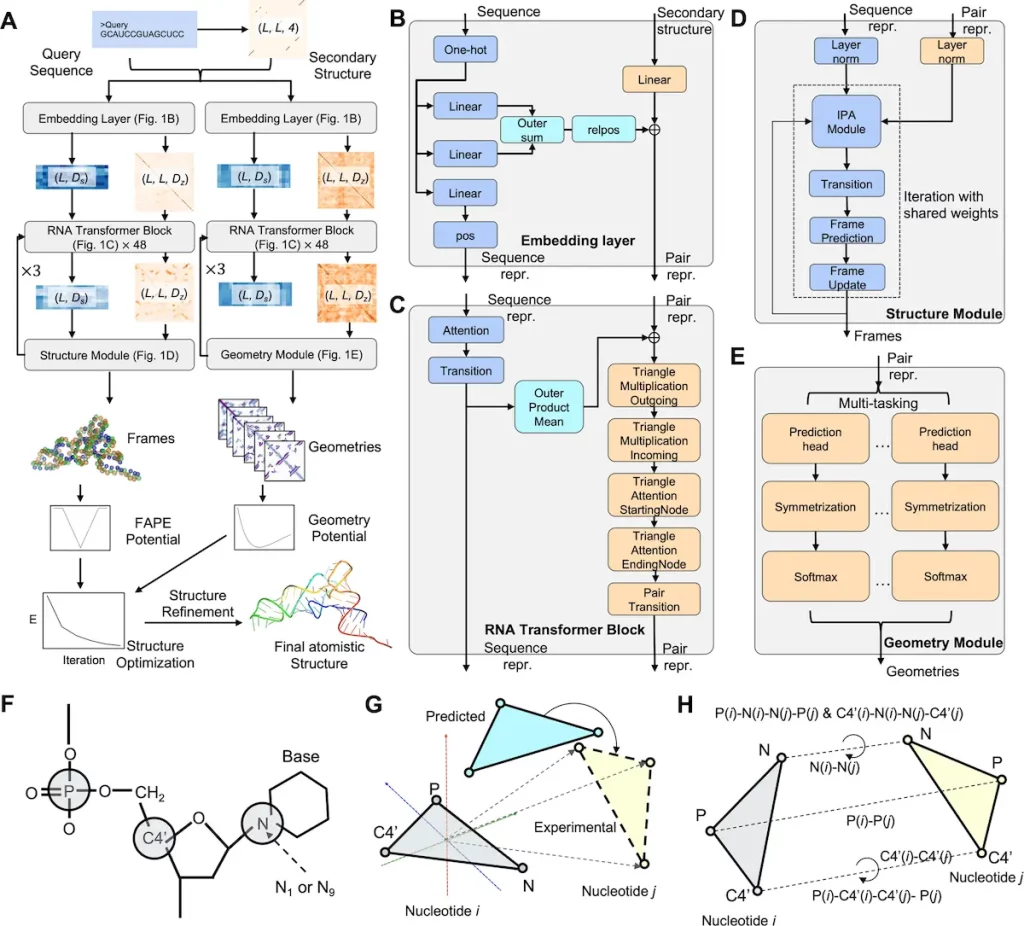
Image Source: https://doi.org/10.1038/s41467-023-41303-9
DRfold Shines: Competing with Cutting-Edge Deep Learning Methods and Impressing in Blind RNA Structure Prediction (CASP15)
The new RNA structure prediction method DRfold proves its worth by exceeding existing approaches and outperforming state-of-the-art deep learning models in terms of performance. The method incorporates end-to-end and geometry-based approaches through potential integration, offering flexibility for pipeline growth. Notably, DRfold shows its effectiveness without relying on complicated MSAs by outperforming single sequence-based approaches and competing with MSA-based techniques. DRfold’s ability to include geometry predictions from DeepFoldRNA, leading to even higher accuracy, further exemplifies its versatility. This adaptability establishes DRfold as a promising tool for precise RNA structure prediction and underlines the possibility of further improving it by incorporating additional techniques. DRfold performs excellently in a community-wide assessment (CASP15), highlighting its competitiveness and resilience in blind RNA structure prediction.
Article Source: Reference Paper | The DRfold standalone package is available on the Website and on GitHub.
Learn More:
Prachi is an enthusiastic M.Tech Biotechnology student with a strong passion for merging technology and biology. This journey has propelled her into the captivating realm of Bioinformatics. She aspires to integrate her engineering prowess with a profound interest in biotechnology, aiming to connect academic and real-world knowledge in the field of Bioinformatics.
















