Large Language Models (LLMs) have emerged as dynamic tools with the ability to transform the landscape of biological research in a world driven by data and technology. These advanced AI models, including well-known names like BERT and GPT, are not confined to linguistics but have found their way into the complex realm of biology. In this article, let’s explore the potential of LLMs in addressing pressing biological challenges, such as drug discovery, disease diagnosis, genomics, and more.
What are Large Language Models?
ChatGPT, along with similar models, has gained widespread attention and adoption. Many people even integrate it into their daily routines. What makes ChatGPT and similar models remarkable? These LLMs represent a facet of artificial intelligence (AI) specifically formulated to understand, process, and even create “human-like” text. Developers construct them using transformers—a type of neural network—as their architecture; they pre-train these on copious amounts of textual data to assimilate the patterns, structures, and nuances inherent in human language.
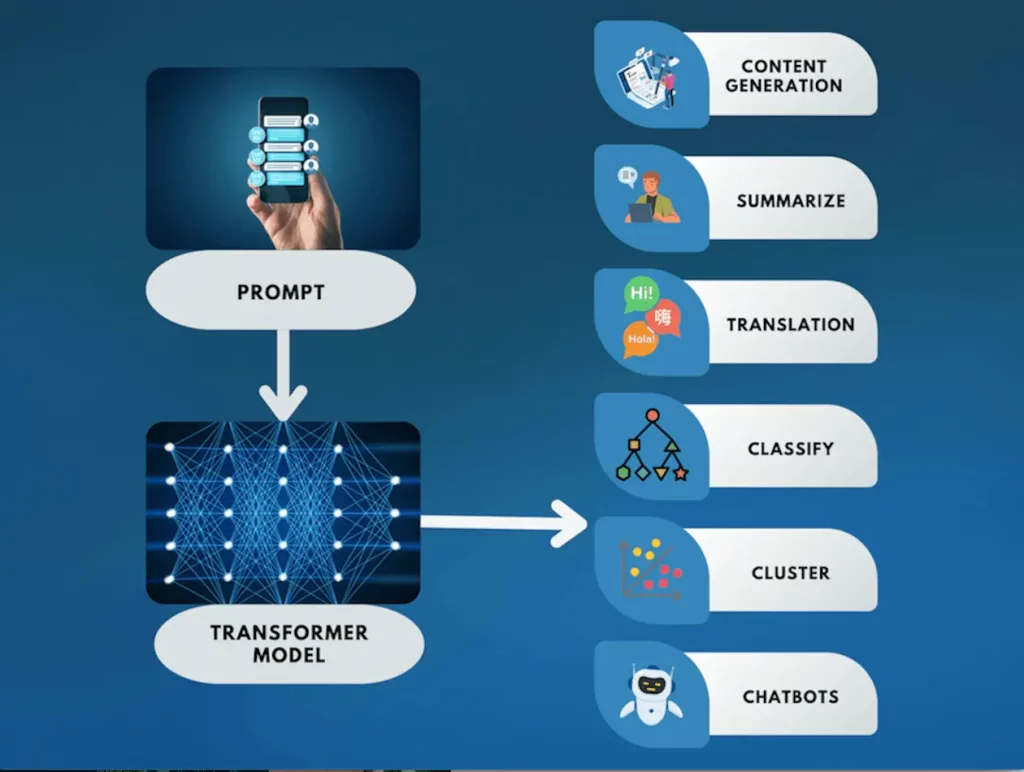
Capable of executing manifold tasks linked with natural language processing (NLP)—including language translation, text summarization, text editing, information extraction, sentiment analysis, and question answering—these AI systems require only a user-provided task description in human terms. Given such directives as input, they generate pertinent outputs tailored to meet the requirements stipulated by the user; their capacity for adaptation ensures that replies remain germane to individual preferences, thus increasing their overall usefulness. Their text processing and generation abilities, which expand across numerous areas, demonstrate remarkable versatility.
Large Language Models offer enthralling marketing content, insightful financial analysis, or efficient customer support through AI bots, further broadening their scope. They use this skill set to assist with evaluating and debugging programming languages. In the field of healthcare, their most fascinating application emerges. At a basic level, LLMs conduct literature reviews, summarize, and explain vast amounts of research papers while forming connections between them.
Advanced-level analysis allows them to dissect large datasets on chemical compounds, predict interactions among these components, and facilitate drug development. They interpret the genes or proteins accountable for certain traits or diseases in living entities by scrutinizing proteomic and genomic data. By examining evolutionary data, they propose innovative theories and potentially unidentified links in evolution’s progression. Even in designing efficient biomedical experiments for improved outcomes in biological research, they can provide assistance.
Understanding Large Language Models
The foundation of LLMs is constructed upon five fundamental building blocks, which collectively contribute to their overarching mechanism, which includes:
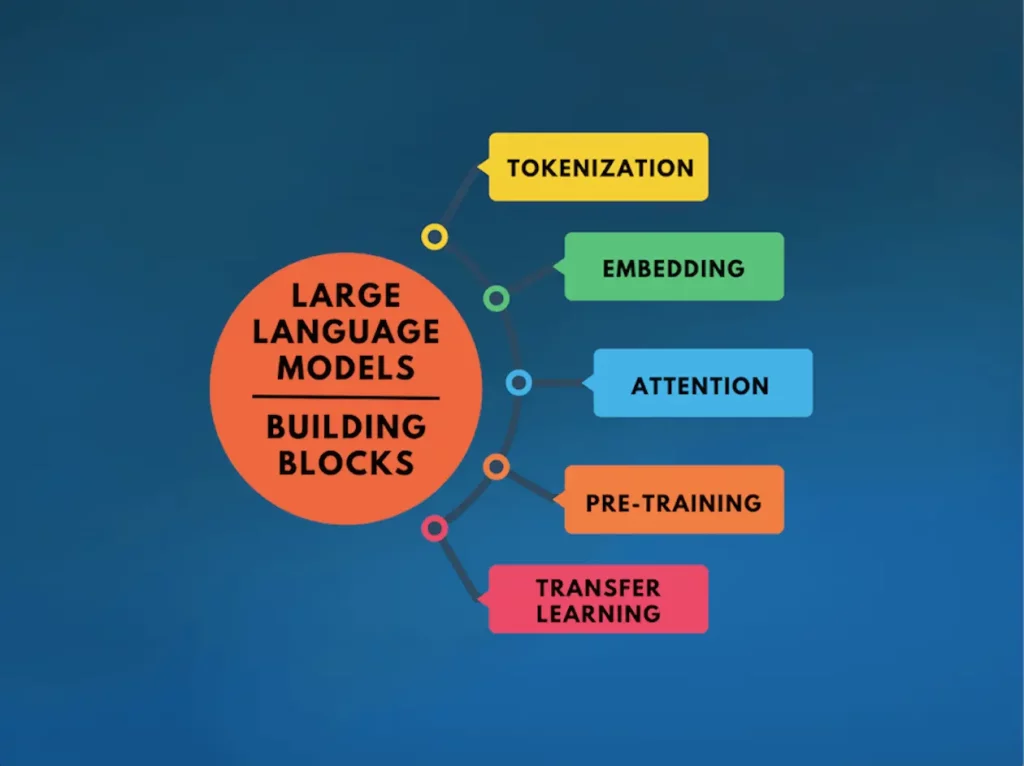
Tokenization: In this process, a sequence of text is broken down into words or tokens by applying algorithms like Byte Pair Encoding (BPE) such that the size of the vocabulary for the model gets limited, but still, the ability to represent any text is retained.
Embedding: It involves the conversion of tokens into vector representations for capturing their semantic meanings such that the neural network can easily process them.
Attention: This process enables the models to attach weights to certain words or phrases depending on the context. This allows the model to be selective and focus on relevant details only.
Pre-training: This process involves training a model on a huge dataset before optimizing it for a specific task so that it can learn all the language patterns and form meaningful associations among the elements of the language.
Transfer Learning: Lastly, the model is optimized for a particular task by utilizing its learnings from the pre-training. This provides the benefit of not using large datasets and extensive training for every new task.
There are two main types of LLMs, differentiated by their underlying transformer architectures. These categories are as follows:
GPT-type Language Models (Decoder-only): GPT stands for Generative Pre-trained Transformer. These are also called autoregressive models. They are trained to predict the next word when given the previous words. Their applications include text generation and question-answering. A few examples of GPT-based models include GPT-3, OPT, and PaLM.
BERT-type Language Models (Encoder-Decoder or Encoder-only): BERT stands for Bidirectional Encoder Representations from Transformers. These models are trained on the Masked Language Model (MLM) paradigm, which involves predicting masked words in the context of surrounding “unmasked” words. Such training allows the model to understand the relationships between the words. These types of models are commonly used for sentiment analysis and named entity recognition. Some examples include BERT, RoBERTa, and T5.
LLMs possess a spectrum of applications that transcend mere text generation, encompassing various domains relevant to biological contexts. Several notable areas within this purview comprise:
Chatbots: LLMs like LaMDA, PaLM 2, Sparrow, ChatGPT, and BlenderBot-3 serve as chatbots by integrating information retrieval, multi-turn interaction, and text generation.
Computer Programming: LLMs like Codex, Polycoder, CodeGen, and AlphaCode are used in programming tasks, such as generating Python functions, translating code between languages, and solving competitive programming questions.
Computational Biology: LLMs are being extensively used in systems biology tasks like protein embedding and genomic analysis. LLM architectures such as ProtT5 and ESM-2 generate protein embeddings from amino-acid sequences or genomic sequences, which are then employed for protein structure prediction, sequence generation, and classification. In the case of genomic analysis, Genome-scale Language Models (GenSLMs) handle long input sequences, pre-trained on gene sequences, and fine-tuned for tasks like identifying new genetic variants. Nucleotide Transformers, trained on nucleotide sequences from different species’ genomes, achieve successful results in genomic prediction tasks, and HyenaDNA, a genomic language model, is proposed to model genomic sequences up to a whopping one million tokens.
The Landscape of Pressing Biological Challenges
The world around us continuously changes, and so do we. With change come opportunities as well as obstacles. As the world progresses, it encounters numerous challenges, from social to economic to even biological. Right now, some of the most pressing biological challenges that humanity is facing are:
- Drug Discovery and Development: With time, pathogens are not only becoming resistant to existing drugs, but also new pathogens are evolving. The drastic lifestyle changes are also rendering many drugs and even our immune systems ineffective. But, discovering and developing new drugs is not only complex but also costly. It takes years to develop effective drugs, which may still fail during clinical trials.
- Disease Diagnosis and Treatment: Many diseases are not fully understood yet; many get detected very late, and some have expensive treatments. All these factors contribute to the patient’s worsening condition or even death. To combat these issues, the research and development of diagnostic tools that can provide comprehension, early detection, and options for affordable alternative treatments are a huge necessity.
- Genomics: Science has come a long way in genomic sequencing, but associating genetic information with clinically effective treatments is still a considerable challenge. Factors such as the complexity of gene interactions, ethical considerations, and the need for large-scale data analysis contribute to this challenge.
- Infectious Disease Control: Medicines are advancing, but so are viruses. Infectious diseases like HIV and malaria continue to maintain their terror, and viruses like Ebola and Zika keep emerging stronger. These viruses are responsible for the loss of lives of millions of people, which necessitates the development of tools for predicting viral outbreaks and evolution along with powerful vaccines and diagnostics.
- Ecological Monitoring and Conservation: The reckless exploitation of the environment, leading to biodiversity loss and ecological imbalances, will bring doom upon ourselves if not checked and controlled. Implementing remote sensing, sensor networks, and big data analysis on a large scale to monitor these environmental damages is still a challenge.
- Agriculture: Urbanization and globalization have immensely contributed to global warming and climate change, leading to food insecurity. Rising temperatures, extreme weather patterns, and irregular changes in precipitation patterns are causing crop failures and disruptions in the food supply chain.
Unveiling the Role of LLMs in Biological Problem-Solving
The integration of LLMs into the field of biology has sparked a transformative paradigm shift. As a testament to their remarkable versatility, LLMs have begun to unravel complex biological enigmas and provide innovative solutions to some of the most intricate challenges in the domain. Several notable “biological” pursuits undertaken by LLMs include:
- Data Analysis and Interpretation: Using LLM as the foundation, researchers from Argonne National Laboratory, the University of Chicago, and others have developed Genome-Scale language models (GenSLMs). Trained on more than 100 million nucleotide sequences, these models can swiftly and accurately analyze the genetic mutations and predict variants of concern in SARS-CoV-2, the virus behind COVID-19.

- Natural Language Processing in Medical Records and Scientific Literature Retrieval: IBM researchers have developed Watson, which utilizes NLP to provide guidance to clinicians in making treatment decisions related to cancer by analyzing patient data. Watson is a cloud-based system that clinicians can access anywhere in the world. It has been tested on real Australian lung cancer cases and has shown immense potential.
- Pattern Recognition in Genomics and Proteomics: ESM-1b is a prime example of an LLM employed in protein structure prediction. It can predict protein structures from amino acid sequences with accuracy.
- Environmental Conservation: Researchers from the University of Oxford, the University of Zurich, and others are integrating climate change data resources so that LLMs can access and present credible information and nuances on climate change.
LLMs Enabling Solutions to Complex Biological Challenges
Let us explore Language Model Models (LLMs) that are actively assisting life science researchers in their endeavors to surmount a diverse array of biological challenges:
LLMs for Biomedical Text Mining
- LLaVA-Med: Researchers at Microsoft have developed a Large Language and Vision Assistant for BioMedicine (LLaVA-Med), which can process text and images. It leverages a dataset extracted from PubMed Central and utilizes the GPT-4 model to self-instruct open-ended biomedical conservations involving pictures and text. It serves as an excellent biomedical virtual assistant.
- PubMedBERT: Developed by Microsoft researchers, this model is trained on abstracts from PubMed and full-text articles from PubMedCentral. It specializes in biomedical text processing and analysis. It has demonstrated efficiency in answering biomedical questions and detecting medicine mentions from tweets.
- BioBERT: Developed by researchers from Korea University, this model specializes in biomedical NLP tasks such as biomedical named entity recognition, relation extraction, and question answering.
- Med-PALM: Developed by researchers from Google, Med-PALM specializes in providing elaborate answers to biomedical questions. It also has a multimodal version that can analyze images as well as text.
- PubMedGPT: Similar to PubMedBERT, this model has been developed by the Stanford Center for Biomedical Informatics Research (BMIR) and specializes in biomedical text mining.
- BioLinkBERT: Based on LinkBERT, this model has been developed by researchers from Stanford University, and it is used for various biomedical NLP tasks.
- DRAGON: Developed by researchers at Stanford University, the self-supervised approach, DRAGON (Deep Bidirectional Language-Knowledge Graph Pretraining), combines text and Knowledge Graph (KG) data, creating joint representations of both modalities. On various downstream tasks like question answering – particularly in complex reasoning and low-resource question answering domains – this method outperforms existing models significantly. It also achieves state-of-the-art results in BioNLP tasks.
- BioGPT: Microsoft researchers have developed BioGPT, a domain-specific generative Transformer language model. The specific aim of this pre-training is to amplify the capabilities of prior models in generating text within the biomedical field rather than merely executing discriminative tasks. BioGPT produces fluent and pertinent descriptions for terms in biomedicine. BioGPT surpasses previous models on most of the six biomedical NLP tasks.
- ChatGSE: Researchers from Heidelberg University have developed ChatGSE, a web application that combines human ingenuity with machine memory to enhance biomedical analyses. It tackles the challenges Large Language Models (LLMs) present, which include a lack of general awareness, logical deficits, and tendencies toward hallucination.
LLMs for Genomic Analysis
- GeneGPT: Researchers from the National Library of Medicine (NLM), National Institutes of Health (NIH), and University of Maryland, College Park, US, developed GeneGPT, which leverages the Web APIs of the National Center for Biotechnology Information (NCBI) to answer questions on genomics. The integration of Web API tools reduces hallucinations in the LLM. GeneGPT has surpassed Bing, ChatGPT, and GPT-3 in terms of performance.
- DNAGPT: Developed by researchers at the Southern University of Science and Technology, Tencent AI Lab, Shenzhen, China, and the City University of Hong Kong, DNAGPT can analyze numerous DNA sequences from various species. It can detect non-coding regions and even analyze artificial human genomes.
LLMs for Drug Development
- DrugGPT: Researchers from Xi’an Jiaotong University have created DrugGPT, which simplifies the drug design process. It effectively overcomes the problem of vast chemical space in the conventional drug design process, increasing the speed and accuracy of that process. Even better is its ability to extrapolate known information onto new scenarios and complete novel tasks.
- CancerGPT: Researchers from the University of Texas and the University of Massachusetts, USA, have introduced CancerGPT, which can accurately predict the outcome of drug combinations on the tissues of cancer patients without requiring extensive data or features.
- SynerGPT: This model has been developed by researchers from the University of Illinois and Allen Institute for Artificial Intelligence. It predicts unseen drug combinations from a minimal amount of data.
Ethical Considerations and Future Prospects
LLMs certainly seem like a revolution, but before their full potential is tapped, we need to be considerate of the ethical issues that may arise. One issue is that of data privacy. Since LLMs collect large amounts of data, it is possible that they may not maintain the privacy of that data, especially when they are tasked with analyzing patient data for medical purposes. Also, when LLMs are trained on internet data, they have a high chance of consuming misinformation as well as biased content. If LLMs provide misleading answers while answering questions, authenticity cannot always be verified, resulting in issues. Probabilistic programming presents a challenge regarding dependable application reliance. Such disparities arise from distinct approaches to solving problems. Noteworthy is the fact that LLMs possess a significant environmental impact due to their large carbon footprint and copious energy consumption.
These disadvantages might prompt the spread of bogus data as valid and lead to issues, particularly while managing delicate information. However, this doesn’t imply that LLMs have no extension to be applied in tackling issues of this present reality. We have to execute mindful and severe morals and guidelines. An extraordinary method for doing so is through interdisciplinary joint effort, i.e., cooperation between man-made intelligence specialists, scholars, clinical experts, ethicists, and policymakers to accomplish the shared objective of combatting the moral issues of LLMs. Such measures can make a future where man-made intelligence permits people to tailor exact customized therapies for each person, novel medications to dispense with malignant growth from the roots, extraordinarily control the spread of viral infections, and secure the eventual fate of the planet.
Conclusion
LLMs play with text and achieve an immense assortment of errands that can be used in different fields with certifiable applications. Biological science is a huge space, and integrating LLMs into science can smooth out different methods in organic exploration as well as extend its extension. By simply investigating printed information, LLMs can fuel exact medication configuration, find novel and reasonable therapy choices for lethal sicknesses like malignant growth, alert about transformations prompting hereditary illnesses, track down ways of halting the spread of irresistible infections, and the rundown is ceaseless. An ever-increasing number of endeavors ought to be placed into defeating the difficulties of completely remembering LLMs for natural exploration, with the goal that an idealistic future in medical care turns into a reality.
Article Source:
- Yang, J., Jin, H., Tang, R., Han, X., Feng, Q., Jiang, H., Yin, B., & Hu, X. (2023). Harnessing the power of LLMs in Practice: A survey on ChatGPT and Beyond. arXiv (Cornell University). https://doi.org/10.48550/arxiv.2304.13712
- Kaddour, J., Harris, J. S., Mozes, M., Bradley, H., R, R., & McHardy, R. (2023). Challenges and applications of large language models. arXiv (Cornell University). https://doi.org/10.48550/arxiv.2307.10169
- Zvyagin, M., Brace, A., Hippe, K., Deng, Y., Zhang, B., Bohorquez, C. O., Clyde, A., Kale, B., Perez-Rivera, D., Ma, H., Mann, C. M., Irvin, M., Pauloski, J. G., Ward, L., Hayot, V., Emani, M., Foreman, S., Xie, Z., Lin, D., . . . Ramanathan, A. (2022). GenSLMs: Genome-scale language models reveal SARS-CoV-2 evolutionary dynamics. bioRxiv (Cold Spring Harbor Laboratory). https://doi.org/10.1101/2022.10.10.511571
- Somashekhar, S P & Sepulveda, martin-j & Norden, Andrew & Rauthan, Amit & Arun, Kumar & Patil, Poonam & Yethadka, Ramya & C., Rohit Kumar. (2017). Early experience with IBM Watson for Oncology (WFO) cognitive computing system for lung and colorectal cancer treatment.. Journal of Clinical Oncology. 35. 8527-8527. https://ascopubs.org/doi/10.1200/JCO.2017.35.15_suppl.8527
- Kraus, M., Bingler, J. A., Leippold, M., Schimanski, T., Senni, C. C., Stammbach, D., Vaghefi, S. A., & Webersinke, N. (2023). Enhancing Large Language Models with Climate Resources. arXiv (Cornell University). https://doi.org/10.48550/arxiv.2304.00116
- Li, C., Wong, C., Zhang, S., Usuyama, N., Liu, H., Yang, J., Naumann, T., Poon, H., & Gao, J. (2023). LLAVA-MeD: Training a large Language-and-Vision assistant for biomedicine in one day. arXiv (Cornell University). https://doi.org/10.48550/arxiv.2306.00890
- Han, Q., Tian, S., & Zhang, J. (2021). A PubMedBERT-based Classifier with Data Augmentation Strategy for Detecting Medication Mentions in Tweets. arXiv (Cornell University). https://doi.org/10.48550/arxiv.2112.02998
- Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2019). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234–1240. https://doi.org/10.1093/bioinformatics/btz682
- Singhal, K., Azizi, S., Tu, T. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023). https://doi.org/10.1038/s41586-023-06291-2
- Stanford CRFM introduces PubMedGPT 2.7B. (n.d.). Stanford HAI. https://hai.stanford.edu/news/stanford-crfm-introduces-pubmedgpt-27b
- Yasunaga, M., Leskovec, J., & Liang, P. (2022). LinkBERT: Pretraining Language Models with Document Links. arXiv (Cornell University). https://doi.org/10.48550/arxiv.2203.15827
- Edwards, C. K., Naik, A., Khot, T., Burke, M. D., Ji, H., & Hope, T. (2023). SynERGPT: In-Context learning for Personalized drug synergy prediction and Drug Design. bioRxiv (Cold Spring Harbor Laboratory). https://doi.org/10.1101/2023.07.06.547759
- Birhane, A., Kasirzadeh, A., Leslie, D. et al. Science in the age of large language models. Nat Rev Phys 5, 277–280 (2023). https://doi.org/10.1038/s42254-023-00581-4
- Harrer, S. (2023). Attention is not all you need: the complicated case of ethically using large language models in healthcare and medicine. EBioMedicine, 90, 104512. https://doi.org/10.1016/j.ebiom.2023.104512
Learn More: