Standford researchers have discoursed the critical aspect for the integration of Foundation Models (FM) in the Healthcare system, that is evaluation of the reliability and extent of applicability of FM models, and advocated metrics, tasks, and datasets for improving the assessment framework that would facilitate effective determination of the value of clinical FMs. The research paper, published in Nature Journal, also presents a well-organized assessment of the architectures, training datasets, capabilities, public accessibility, and limitations of 84 different “Clinical FMs’ which were formulated on the basis of electronic medical record (EMR) data.
A Glimpse into Relevance of the Research
The breakthroughs and popularity of Artificial Intelligence have incited inquisitiveness regarding whether reliable and accurate clinical advice can be generated using such models. Multiple projects were conducted over the past decade to check the usefulness of Machine Learning models (ML) in the medical field, and many of them declared positive outputs. Since the basis for evaluating the clinical practicability of such models is still not reasonably established, considering the critical concern for public health associated with it, the authors inspect the justification of the hype oriented toward the aspiration of AI’s mass incorporation in clinical settings.
Clinical Foundation Models offer advantages over conventional ML, so the study incorporates a review of 84 different clinical FMs built from electronic medical record (EMR) data, including structured (e.g., billing codes, demographics, lab values, and medications) and unstructured (e.g., progress notes, radiology reports, and other clinical text) EMR data, but excluded images, genetics, and wearables to manage the scope of this review. Therefore, a common suspicion is whether these Clinical Foundation Models overrate their effectiveness and fabricate exaggerated promises or can actually offer genuine decisions when the matter is related to precious life; is officially evaluated through this endeavor.
Clinical Foundation Models: Advantages Over Traditional Machine Learning Models
Foundation models (FM), in contrast to the conventional Machine Learning models, are “pre-trained with prodigious unlabeled data, accommodates vast parameters, representations, and provides a foundation for various downstream tasks without much requirement for adaptation or retraining. One such prominent example of FM is ChatGPT, developed by OpenAI. Utilizing electronic medical record data (EMR), many Clinical FMs have also been formulated. FMs have changed the trajectory of ML development. Unlike conventional ML models, the researchers defined advantageous attributes of Clinical FM models.
- Clinical FMs offer more sensitive and specific predictive performance than traditional MLs.
- Clinical FMs don’t require task-specific labeled data, and often ‘zero-shot’ or ‘few-shot’ learning is sufficient for their performance.
- FMs can ingest a wide range of data modalities, such as structured codes, lab values, clinical text, images, speech patterns, etc., as inputs and incorporate them into a single, unified representation. For instance, a model might simultaneously consider an MRI scan and progress notes while predicting a patient’s optimal treatment.
- Easy deployment of the Clinical FMs.
- Clinical FMs models can potentially be applied in new clinical applications, such as writing improved and coherent insurance appeals.
- Importantly, convenient interaction can be achieved between the models and users in natural language via prompting.
“A Shaky Foundation”?: Reviewing the Current Status of Clinical Foundation Models
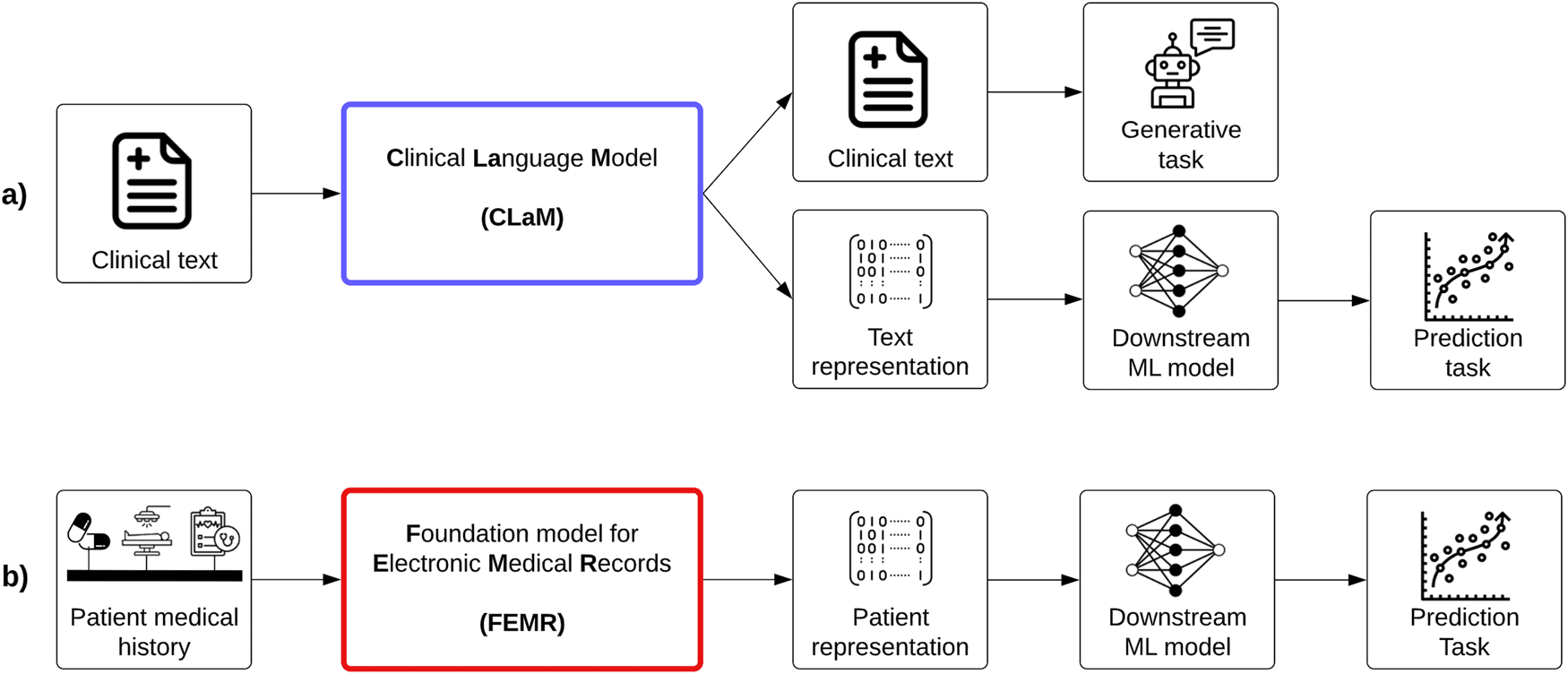
The authors arranged Clinical FMs into two categories; one is Clinical language models (CLaMs), which are a subtype of large language models (LLMs), which could extract prescribed drug names from a physician’s notes, interpret and summarize medical dialogues and also reply to patient queries.
CLaMs are trained with either or both biomedical texts, such as publications available in PubMed and clinical texts acquired from the MIMIC-III database, which includes approximately a million notes written between 2001 and 2012 in the ICU of the Beth Israel Deaconess Medical Center. Therefore, the researchers cognized a significant gap since medical information outside of the timeframe – after 2012 and these two databases- PubMed and MIMIC-III; aren’t incorporated in the CLaMs models.
Moreover, the journal outlined a surprising issue that most of the CLaMs were not validated on clinical text but rather evaluated on traditional NLP-style tasks such as named entity recognition, relation extraction, and document classification on either MIMIC-III (clinical text) or PubMed (biomedical texts).
The clinical text has its own unique structure, grammar, abbreviations, terminology, formatting, and other peculiarities not found in other domains, and good performance on a clinical NLP task does not provide satisfactory evidence in a hospital scenario; therefore, it is most likely that those models may be overestimating their expected performance in a practical setting.
Not only that, the researchers pointed out that most CLaMs only evidenced only one out of six value propositions, “Benefits of Clinical FMs.” Basically, it can’t be enunciated with an assurance that “Model A achieves high precision on named entity recognition on 2,000 discharge notes from MIMIC-III” is very different than “Model A should be deployed across all of Health System X to identify patients at risk of suicide.”
The other category is Foundation models for EMRs (FEMRs), which are trained with the patient’s medical history, such as structured codes, lab values, claims, and clinical text. FEMR generates a machine-understandable “representation” of that patient as an output, termed “patient embedding,”; a high dimensional vector accommodating large amounts of patient information. FEMR can generate medically relevant tasks.
Herein, the dependence on structured codes constrains FEMR’s generalizability and inconsistencies, which result in an unclarified representation of the patient’s state. Apart from that, open-sourcing pre-trained FEMRs is also a challenge. Moreover, there were no common grounds for assessing the performance of FEMRs across different publications. FEMRs also fail to meet the six primary value propositions of clinical FMs listed in “Benefits of Clinical FMs.”
Amending the Evaluation Parameters of Clinical FMs
Pointing out the major deficiencies, the researchers recommended some alternative strategies to comprehend the clinical value of the models.
- Instead of showcasing predictive performance through classification and regression tasks based on AUROC, AUPRC, and F1 Score, employing ranking-based metrics can potentially improve the basis of evaluation.
- The ability of the model to exhibit performance by decreasing dependency on labeled data is also an important parameter.
- Evaluating how less resourceful the model is in terms of cost of hardware, computer or memory, and time.
- The capabilities of conducting ‘emergent’ tasks previously unthinkable to perform.
- Demonstration of multimodal reasoning in an evaluation task.
- Improvements in human-AI interfaces.
Conclusion
Inspired by the previous approaches to evaluating large language models (LLMs) comprehensively, the study aspires to initiate the discussion of how FMs should be assessed to eradicate the chances of overestimating prior to applying for clinical purposes. As Clinical FMs offer advantages over conventional ML models, the evaluation basis should also be based on how well the models excel in those advantageous aspects. The study reveals the impractical background of clinical FMs compared to a real-life scenario, but the proposals certainly will provide a strong foundation for bridging these concerning gaps for future developments.
Article Source: Reference Paper
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.