Motivated by the success of the GPT (Generative Pre-trained Transformer) model, researchers from the Southern University of Science and Technology, Tencent AI Lab, Shenzhen, China, and the City University of Hong Kong have developed DNAGPT, a generalized foundation model capable of simultaneously processing multiple DNA sequences from various species. Its unique token design allows users to optimize prompts per their requirements, making it useful in any task. The researchers found that the model improves with pre-training, and it was evaluated on different jobs, including classification (grouping DNA sequences into categories), regression (predicting numerical values), and generation (creating new DNA sequences).
The Pursuit of Building a Universal Tool for DNA Sequence Analysis
DNA sequences, specifically the large non-coding regions, contain vital, undiscovered information which, when explored, can reveal novel insights underpinning the various mechanisms of life. Different species have similarities along with divergences in their genetic sequences. Therefore, building a generalized tool that can analyze DNA sequences from many different species is imperative.
The recent advent of foundation models has boosted natural language understanding to a great extent, but DNA sequences have added layers of complexity and diversity by virtue of the sequences themselves, expression levels, etc. Earlier, an attempt was made for a generalized analysis of DNA sequences across species. A model called DNABERT was developed to obtain generalized learning from various sequences. It was pre-trained only on the human reference genome.
While this model was successful in dealing with tasks related to genomic signals and regions (GSR), it failed at other tasks, such as determining the correlation between DNA sequences and mRNA abundances and the generation of pseudogenes for research purposes. Therefore, scientist Daoan Zhang and his team have developed DNAGPT, a generalized foundation model pre-trained on 10 billion base pairs from nine different species (Arabidopsis thaliana, Caenorhabditis elegans, Bos taurus, Danio rerio, Drosophila melanogaster, Escherichia coli, Homo sapiens, Mus musculus, and Saccharomyces cerevisiae).
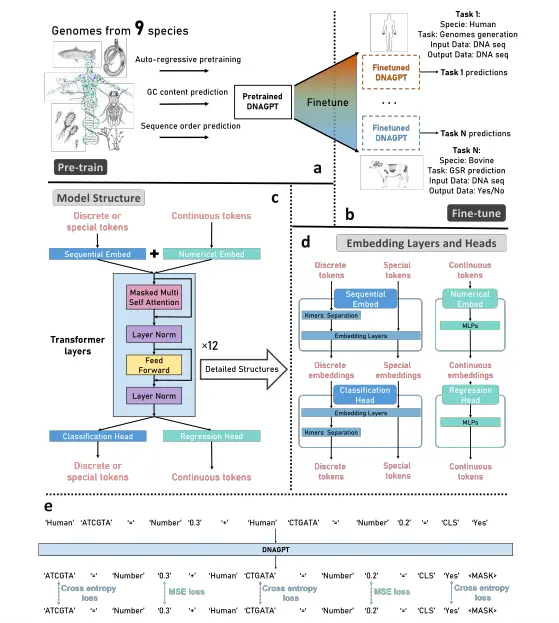
During training, DNAGPT learns not only to predict the next base in a sequence (auto-regression task) but also to predict other characteristics of DNA, such as the content of certain base pairs (Guanine-Cytosine) and the order of the sequence. To make DNAGPT useful for different DNA analysis tasks, a symbolic language is created. This language allows different tasks to be encoded into a sequence format so that the model can handle and switch smoothly between various downstream tasks without needing separate training for each task.
The Brilliant Architecture of DNAGPT Revealed!
The architecture of DNAGPT utilizes a type of artificial intelligence called a transformer. The model has been trained using transformer-based auto-regressive decoding with masked self-attention. The masked self-attention module restricts the attention of the model to only the past tokens. To make the model handle numerical information effortlessly, it was pre-trained on both DNA sequences as well as numbers. The model effectively deals with both discrete tokens (encoded DNA sequences) and continuous tokens (encoded numbers).
The encoding of DNA sequence involves the division of the sample DNA sequence into smaller, non-overlapping units called k-mers. These k-mers are then sent to the Sequential Embedding Layer to be encoded or transformed into embeddings (numerical representations of the DNA sequence). The numbers are also encoded as embeddings using a separate layer called the Numerical Embedding Layer. Then, both these embeddings are combined and sent into a GPT model.
The GPT model processes the combined embeddings and generates the outputs. The outputs are further divided into two types. The Classification Head processes the embeddings of the discrete tokens to classify different tokens, and the Regression Head processes the embeddings of the continuous tokens to generate numbers.
The problem with most of the existing models is that they treat DNA sequential information like regular natural language without taking into account its unique properties. Unlike natural languages, a DNA sequence does not have a defined structure like words, sentences, or paragraphs. Treating it like natural language creates a significant difference between the training process and the actual tasks the model needs to perform.
To eliminate such problems, the symbolic language architecture is employed in DNAGPT. This architecture considers individual base pairs as the smallest units in DNA sequences and utilizes non-overlapping k-mers to generate “DNA words.” The “DNA words” are combined to form “DNA sentences,” which in turn are combined to form “DNA paragraphs.”
This “DNA language” consists of different types of tokens apart from discrete and continuous tokens, which include:
- Instruction tokens provide instructions regarding what sequence of tokens should be generated next.
- Connection tokens establish relationships between different tokens. For example, the ‘+’ symbol represents combining two series of tokens.
- Classification tokens represent various classes, categories, or labels to be assigned to specific tokens.
- Regression tokens indicate that the model should generate numeric values based on continuous token embeddings.
- Reserved tokens include ‘0’ to ‘9’, unused capital letters like ‘K’ and ‘L,’ and special symbols. They are used for very specific tasks.
To obtain the expression level of an mRNA from the DNA sequence and the mRNA half-life value, the input can be encoded as ‘Human’’ATGTC’’+’’M-0.3484’’=’’M’’0.9854’. This means that we want the model to generate information from the human DNA sequence ATCGTC and input the number ‘-0.3484’ to output the number 0.9854. ‘M’ indicates that the numbers in front of it be encoded into the model.
Behind the Scenes: Casting Light on the Training of DNAGPT
Two versions of DNAGPT, DNAGPT-H, and DNAGPT-M, were pre-trained. The former was trained on the human reference genome while the latter on DNA sequences from nine species to get a grasp on performing three tasks for superior DNA sequence analysis.
- Next Token Prediction:- DNAGPT was trained by adding more parameters and a lot of training data so that it can perform the next token prediction in the context of DNA sequences. Next token prediction is a technique through which the next possible token is determined based on the previous tokens.
- Guanine-Cytosine (GC) Content Prediction:- GC content can provide important information related to genome structure like structural variation, transcriptional activity, etc. Hence, to enable DNAGPT to predict GC content efficiently, GC content was represented as a number and fed as input into DNAGPT. This allows DNAGPT to be trained on numbers as well as sequences, making it adaptable to diverse data types. Also, the GC content prediction task was inserted at random places in the input paragraph to make it acquainted with different downstream tasks.
- Sequence Order Prediction:- DNA sequence order holds importance in gene expression and transcription. Certain signals, such as the TATA box, have specific positions that guide the expression and transcription processes. Hence, a sequence order prediction task was designed, which involved the random reversal of DNA sequences to assess whether the model could predict the reversal or not. GPT models have a feature called unidirectional attention, which means they can only infer and create tokens from left to right. Thus, they do not rely on the forward sequence for inferring tokens, rather, they predict from a global perspective.
To train DNAGPT effectively, various loss functions were implemented to calculate the error between the model’s predictions and the expected answers. For the next token prediction and sequence order prediction, a cross-entropy loss function was used. This loss function measures the difference between the probabilities assigned by the model and the true probabilities of the correct answers. For GC ratio prediction, a different loss function called mean squared error (MSE) loss was used, which calculates the average squared difference between the model’s predicted GC ratio and the true GC ratio.
Taking a Look at the Outstanding Features of DNAGPT
- Recognition of Genomic Signal Regions (GSRs): DNAGPT is capable of recognizing various genomic signals and regions (GSRs) from DNA sequences. It has been specifically assessed on the recognition of polyadenylation signals (PAS) and translation initiation sites (TIS) in different organisms, including humans, mice, bovines, and fruit flies.
- Improved Performance: DNAGPT consistently outperforms previous state-of-the-art methods, such as GSRNET and DeepGSR, in the recognition of GSRs. It achieves higher accuracy and better results in most cases, surpassing both modified DNABERT and DNABERT-H models.
- Generalization Across Species: DNAGPT, especially DNAGPT-M, which is pre-trained on DNA sequences from multiple species, demonstrates performance improvements not only in non-human organisms (mouse, fruit fly, and bovine) but also in human GSR recognition tasks. This suggests that incorporating DNA from other species enhances the understanding of human DNA.
- Attention Mechanism: DNAGPT utilizes an attention mechanism to identify GSRs. Attention maps generated by DNAGPT-M reveal that the model focuses on non-coding regions to identify TIS and PAS accurately. The PAS signal, located after the PAS sequence, provides a prompt to the model, facilitating the extraction of task-relevant information from the subsequent sequence.
- Recognition of Non-Coding Regions: DNAGPT demonstrates that non-coding regions play a crucial role in GSR recognition. Attention maps show that DNAGPT-M has a broader attention range for PAS recognition, as the PAS is situated in the middle of non-coding regions. Similarly, for TIS recognition, DNAGPT-M focuses its attention on non-coding regions located before the coding region.
- Analysis of Artificial Human Genomes: DNAGPT can generate pseudo-human genomes, which can protect genetic privacy and reduce the cost of genetic sample collection. Evaluation of DNAGPT-generated genomes through principal component analysis (PCA), allele frequency analysis, and linkage disequilibrium analysis shows that DNAGPT-H and DNAGPT-M fit the distribution of real human genomes more accurately compared to other models such as GAN and RBM.
- Adjustable Generation Temperature: DNAGPT allows control over the randomness of the generated DNA sequences by adjusting the generation temperature. Higher temperatures increase the diversity of generated sequences while maintaining correlations with real distributions. Users can adjust the temperature to control the diversity and authenticity of the generated sequences.
- mRNA Expression Prediction: DNAGPT can extract abundant information from DNA sequences and predict mRNA expression levels of genes directly from genomic sequence information. It outperforms Xpresso in predicting mRNA abundance in both human and mouse species, with DNAGPT-M showing improved results for mouse species.
Conclusion
There is immense genetic information hidden in our DNA sequences. Comprehending, analyzing, and interpreting vast genetic information is the secret to solving many intricate mysteries of life. DNAGPT serves as a simplified model that can process multiple DNA sequences efficiently, revealing many important aspects of genes that are imperative to understanding life processes in great detail across several species. It provides an all-in-one solution and a novel perspective on the many endeavors of genomic research.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.










