A collaborative effort between the Departments of Pharmacology, Chemistry, and Biochemistry at the University of California, San Diego, led to the development of MitoSpace, a self-supervised deep learning model. It can be considered an ‘atlas’ to map various phenotypic characteristics of the mitochondria’s reactions and responses to different drugs. It is a high-content image-based screening process with the goal of recognizing substances that can alter cellular phenotypes. The deep learning element adds a twist to this process by creating a ‘latent space’ from the images fed to it without requiring any data labels to train it.
Traditional approaches for phenotypic profiling
Traditional methods identified compounds that had the ability to reposition disease phenotypes toward healthier phenotypes. The main goal of these methods was to determine the relationship between the morphology of the cell and its state and to find drugs that could shift the cell’s state to a more desired form. Knowledge of the cell’s morphology gives more information on the disease, which in turn is useful to find the corresponding drug that could be used to ‘correct’ the diseased cell’s state.
A drawback of these methods is that they have been unable to cover the vast expanse of the spectrum of potential responses that different phenotypes can exhibit.
A defined set of features, such as the texture, shape, and size of the organelles under study, is prepared and further processed to find a subset of features that classify phenotypes into healthy or diseased types. An example of one of these methods is the NCI-60 cell line panel. To obtain information on the drug’s anticancer properties and link its mechanism of action to potential causative genes and diseases, 60 cell lines from different types of tumors were used.
New methods for screening images
Convoluted neural networks (CNNs) are enhanced methods that extract information from images to a greater extent than traditional methods of image screening. Human-curated feature lists and human annotations are both used to train supervised algorithms.
In contrast, self-supervised deep learning methods do not require any human input. They have had applications in microscopy (largely used in MitoSpace), medical imaging, and predicting protein structures. These methods enable the modeling of distributions of highly complex data.
Introducing MitoSpace!
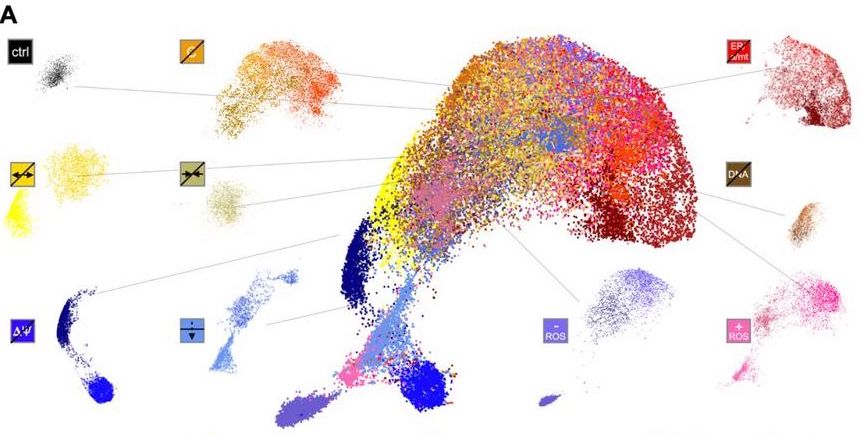
MitoSpace is based on a novel mechanism of ‘cell perceiving’ in contrast to traditionally used ‘cell profiling’ methods. Cell perceiving does not require the data to be labeled and does not require features to be defined beforehand. It involves the association of images with their corresponding drug treatment methods; this information is not given to the model by humans. It is trained to learn similarities and differences between images using associations between given data. The steps that were involved in feature selection and extraction are eliminated. This model also gets rid of any possibility of human bias and can map any variation in data without requiring human assistance.
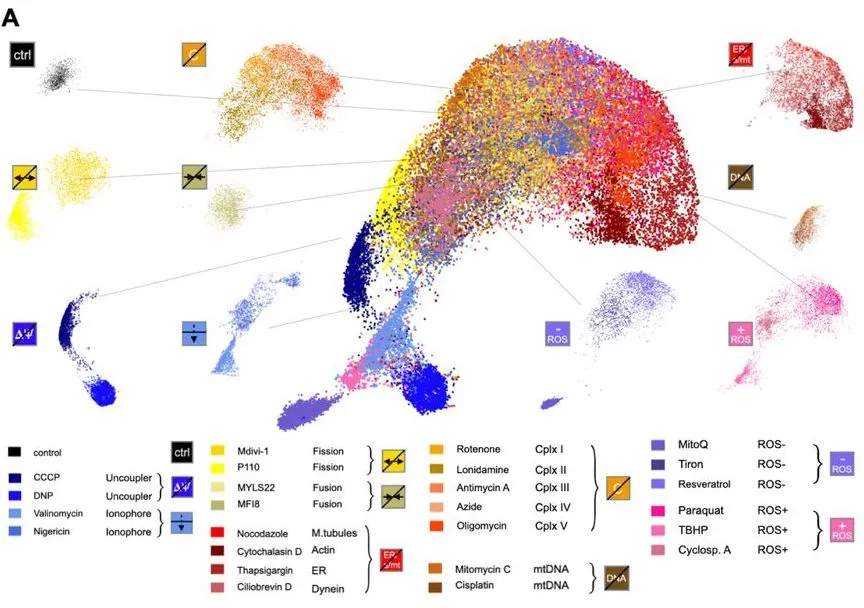
Image Source: https://doi.org/10.1101/2023.09.13.557636
A brief overview of the mitochondria
The network of the mitochondria is diverse, and the organelle itself is vital for cell health. It constitutes 90% of the cell’s energy sources. Conversely, it is also the root cause of a few diseases, like epilepsy, cancer, metabolic disorders, and neurodegenerative diseases. Therefore, understanding the morphology of the mitochondria is key to uncovering the secrets of these diseases and understanding how they can be treated more efficiently. The morphology of the mitochondria is complicated, and manually mapping the mitochondrial network increases the chances of missing out on important features.
Working of the Model
The model uses a two-fold self-supervised deep learning approach: BYOL, a bootstrapping-based deep learning method, and SimCLR, a contrastive method. Two cell lines were used: HeLa cervical cancer cells and Cal27 head and neck squamous carcinoma cells. Eight different classes of drugs were used in this study.
When any sort of stress is applied to the mitochondria, changes are induced in their morphology. The three-dimensional network of the mitochondria changes with time through motility, fission, and fusion processes.
The model creates a ‘latent space,’ basically a map, that comprehends different mitochondrial phenotypes. It can detect changes induced by drugs in the morphology of the mitochondria. The basic principle is to locate phenotypes that are similar within the same latent space. The model learns these similarities through associative mechanisms. Drugs with similar effects on the mitochondria are grouped together. The model shows phenotypic changes that happen from fragmented to tubular forms, as well as extra dimensions that make these changes more noticeable. These findings could not be made with older image screening methods.
Medical applications
- Characterization of drugs: Determining which drug affects which portion of the mitochondria and its extensive network is a costly and laborious process. MitoSpace can be used to perform the same task using microscopy images of cells treated with novel drug candidates.
- Drug discovery: MitoSpace defines a region of interest within the latent space. Images located within this space would have pharmacological characteristics desirable for the drug in question.
- Diagnostics: It is challenging to characterize mitochondrial diseases, as multiple steps are involved in doing so. MitoSpace only requires the user to insert microscopy images of the patient’s cells into the model, where the in-built latent space will contain images that are most similar to the suspected medical condition of the patient. This facilitates screening procedures with more specificity and accuracy.
Conclusion
MitoSpace still has room for further improvement despite its robust and efficient nature. Incorporating a larger variety of drugs, cell lines, and organelles would increase its prediction accuracy. Currently, the model uses 25 drugs as a baseline.
It has the potential to unravel a wider range of effects on the morphology and functions of whatever organelle is under study. Diversification of cell line sets would be beneficial, as different cell lines show different reactions when treated with the same drug. It is important to understand these variations to enable better diagnosis of medical conditions and diseases. Due to their morphological complexities, there is also potential for expanding the model to study organelles other than the mitochondria, such as the cytoskeleton and endoplasmic reticulum. The model currently learns only two-dimensional images, but expanding its application by integrating three-dimensional structures (the true forms of organelles) can enable the model to make more accurate predictions and better understand the hidden secrets in the cell.
Article source: Reference Paper
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Swasti is a scientific writing intern at CBIRT with a passion for research and development. She is pursuing BTech in Biotechnology from Vellore Institute of Technology, Vellore. Her interests deeply lie in exploring the rapidly growing and integrated sectors of bioinformatics, cancer informatics, and computational biology, with a special emphasis on cancer biology and immunological studies. She aims to introduce and invest the readers of her articles to the exciting developments bioinformatics has to offer in biological research today.