Deep-SMOLM is a deep-learning-based estimator created by Washington University in St. Louis researchers that blends machine learning, physical laws, and biological principles. At a rate ten times quicker than iterative estimators, Deep-SMOLM correctly and precisely reconstructs 5D information of simulated biological fibers and experimental amyloid fibrils from images, including highly overlapping single molecule structures.
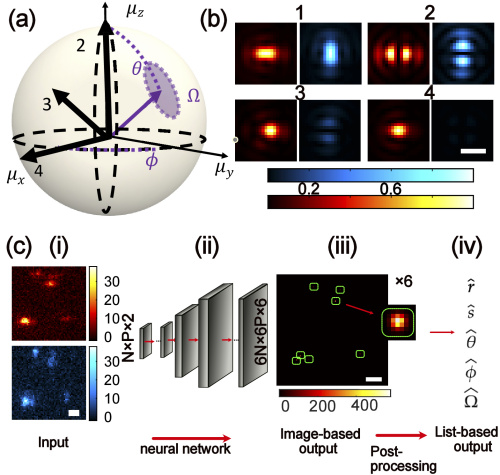
Image Source: https://opg.optica.org/oe/fulltext.cfm?uri=oe-30-20-36761&id=505938###
A series of potent imaging methods known as single-molecule orientation localization microscopy (SMOLM) can scan biological structures at the molecular level and significantly improve spatial resolution over conventional, diffraction-limited microscopy methods. In SMOLM, specific fluorescent molecules are computationally localized from diffraction-limited image sequences and used to produce super-resolution images, time series of super-resolution images, or molecular trajectories. Studies have used SMOLM to visualize amyloid aggregates, actin filaments, and lipid bilayer dynamics.
The study conducted under the supervision of associate professor Matthew Lew has developed Deep-SMOLM. It determines the orientation of a molecule in 3D space and its position in 2D, thereby combining five parameters from a single, noisy, pixelated image with greater precision and speed when compared to iterative estimators. Deep-SMOLM predicts live simulations and experimental images of amyloid fibrils with great accuracy, making it the first deep-learning estimator capable of estimating 5D information from overlapping images of a single molecule.
The research was published in the Optics Express journal.
Major Findings
First, Deep-SMOLM’s capacity to evaluate overlapping images with two molecules separated by various distances (0-1000 nm), with constant and random orientations. As long as the two emitters are separated by at least 139 nm, or an average 43% area overlap, Deep-SMOLM can attain a Jaccard index >0.95 (where the Jaccard index is defined as the size of the overlap divided by the total size of the union of two label sets). More impressively, when processing images of emitters separated by only 414 nm, or 17% overlap, Deep-SMOLM achieves accuracy and precision on par with non-overlapping emitters.
To validate the estimators’ ability to image dense and complex structures, 5D imaging of a 1D fiber was performed. Deep-SMOLM detects and localizes each emitter with excellent detection efficiency, achieving a 0.84 Jaccard index near the middle of the structure where fibers are more separated versus a 0.77 Jaccard index near the poles where overlaps are more prevalent. Deep-SMOLM could not resolve fibers with significant overlaps, i.e., less than 10 nm apart.
Amyloid aggregates are implicated in several neurodegenerative disorders, such as Parkinson’s and Alzheimer’s. Previous research has demonstrated that Nile red (NR) staining, upon transiently attaching to amyloid fibrils, orients itself parallel to their backbones. This enabled the authors to validate the performance of Deep-SMOLM for 5D imaging of orientations and locations from experimental data using imaging NR.
To validate Deep-SMOLM’s performance for analyzing images containing overlaps. Nile red was used at high concentrations to validate Deep-SMOLM’s performance for analyzing images containing overlapping orientations. The deep learning model reconstructed localization microscopy images of intertwined amyloid fibrils spaced 55nm apart with superior detail.
Final Thoughts
According to Tingting Wu, the Ph.D. student and first author of the article, many individuals use AI end-to-end. She split the problem into two steps to reduce the workload on the algorithm and increase its robustness.
The imaging of single molecules in the Lew lab is typically quite “noisy,” containing “specks” or fluctuations that might obstruct an image. Lew stated that coping with that kind of noise can be quite challenging to train for the majority of machine learning neural networks. However, the combination of noise signals and signals from the molecules of interest in microscope images is already known to humans.
Here, the authors have demonstrated how to use Deep-SMOLM, a deep learning-based estimator, to estimate both the 3D orientations and 2D positions of individual molecules from a microscope. Deep-SMOLM achieves superior estimation precision for both 3D orientation and 2D position, which is, on average, within 3% of the best-possible accuracy compared to typical optimization approaches.
As demonstrated on both simulated structures and actual amyloid fibrils, Deep-SMOLM exhibits good performance for predicting overlaps. This capability allows the estimator to achieve a ~2× speed-up in data collection by enabling fluorescent probes to blink at greater rates and to be employed at more significant concentrations. Furthermore, Deep-SMOLM only needs a small amount of training time and data. Compared to other algorithms like RoSE-O, Deep-SMOLM calculates 3D orientations and 2D locations 10 times faster after training.
Way forward, the authors intend to extend Deep-SMOLM to estimate the 3D positions and 3D orientations of overlapping molecules. This method may unlock the potential of SMOLM for cellular and tissue-scale imaging, which may be essential for ensuring that networks are sufficient for in vivo super-resolution imaging.
Ultimately, according to Wu et al., this approach will aid in a better understanding of microscopic biological processes, such as how amyloid proteins aggregate together to produce tangles that are linked to Alzheimer’s disease.
Story Sources: Reference Paper | Reference Article | Deep-SMOLM
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Shwetha is a consulting scientific content writing intern at CBIRT. She has completed her Master’s in biotechnology at the Indian Institute of Technology, Hyderabad, with nearly two years of research experience in cellular biology and cell signaling. She is passionate about science communication, cancer biology, and everything that strikes her curiosity!