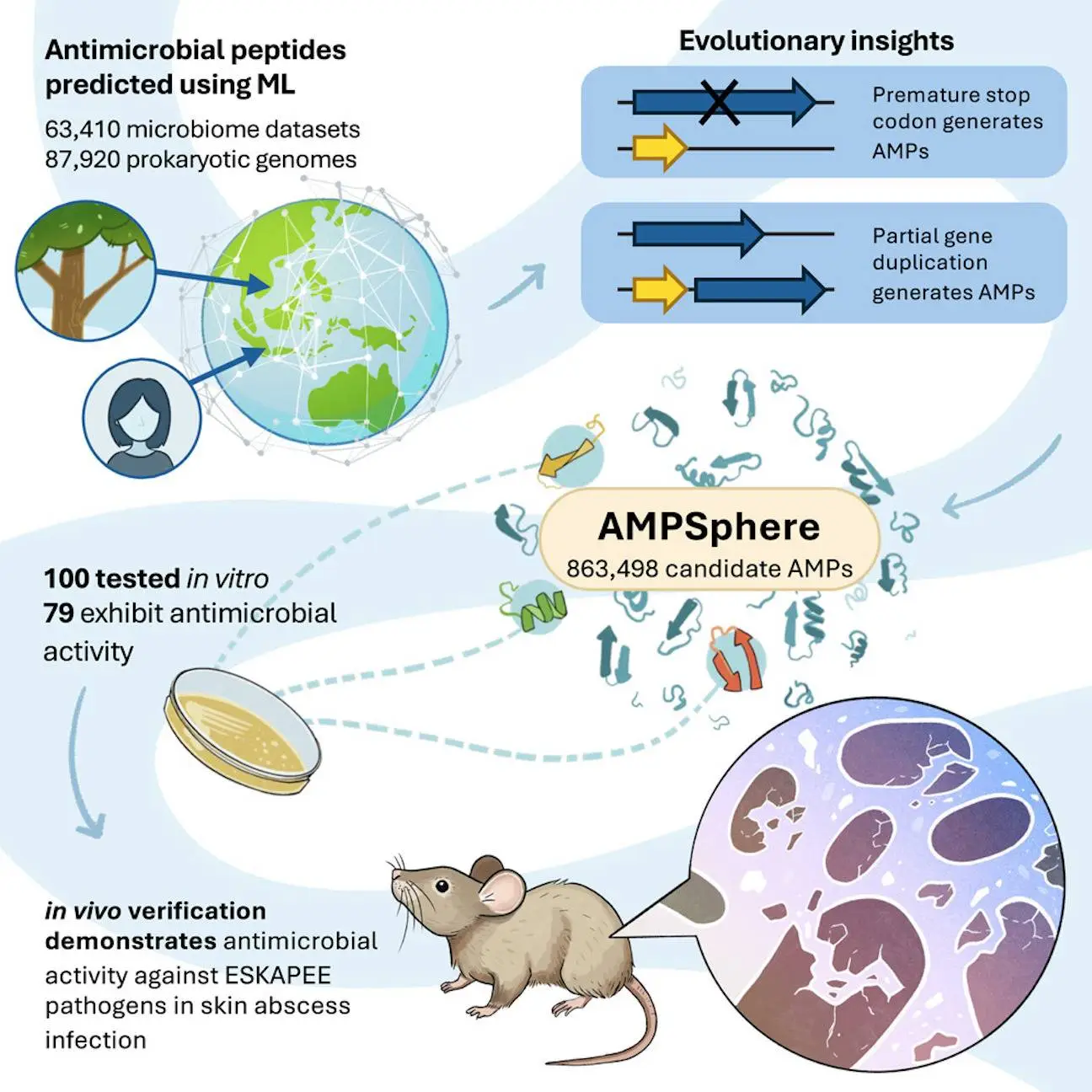
Antibiotic resistance must be addressed immediately with novel antibiotics. By using a large dataset of 63,410 metagenomes and 87,920 prokaryotic genomes from environmental and host-associated habitats, researchers from the University of Pennsylvania and Fudan University have created AMPSphere, a comprehensive catalog consisting of 863,498 non-redundant peptides, few of which match existing databases. The researchers present a machine-learning-based approach to predict antimicrobial peptides (AMPs) within the global microbiome. AMPSphere investigates the evolutionary history of peptides, including the shortening of larger sequences in genes. It shows that different habitats produce different amounts of AMP. The researchers found 79 active peptides using 100 AMPs against human gut commensals and drug-resistant pathogens, of which 63 target pathogens. These active peptides were an accessible resource for the discovery of antibiotics because they showed antibacterial activity by rupturing bacterial membranes.
Introduction
It’s getting more and more challenging to treat antibiotic-resistant infections using traditional medicine. Indeed, 1.27 million individuals die from these illnesses annually. Thus, the development of innovative techniques for antibiotic discovery is desperately needed. Antimicrobial peptides (AMPs) are among the novel antibiotics that researchers can find more quickly, thanks to computational methods. Proteome mining techniques have been employed recently to detect antimicrobial compounds in extinct creatures, with the goal of broadening the range of recognized antimicrobials.
About Antimicrobial Peptides (AMPs)
Antimicrobial Peptides (AMPs) are short sequences that can disrupt microbial growth and are present in all domains of life. They are operationally defined as 10–100 amino acid residues. Most frequently, AMPs disrupt the integrity of cell walls and result in cell lysis. Natural AMPs can be produced by non-ribosomal synthesis, proteolysis, or—the subject of this study—by being encoded in the genome. In their natural environments, pathogens maintain a complex equilibrium between mutualism and antagonism, with AMPs being essential in regulating relationships. Certain strains, such as those of Listeria spp., Vibrio cholerae, Staphylococcus spp., and Shigella spp., drive out rivals and fill their niche.
AMPs have already been utilized therapeutically as antiviral drugs (e.g., telaprevir and enfuvirtide) and show promise as prospective treatments. Clinical trials are currently being conducted on immunomodulatory AMPs and peptides (e.g., pexiganan, LL-37, and PAC-113) that may be utilized to treat bacterial and yeast infections. While the majority of AMPs exhibit broad-spectrum activity, some are limited to their activity against members of the same species or genus that are closely related. Compared to traditional broad-spectrum antibiotics, such AMPs are more focused medicines. Moreover, in contrast to conventional antibiotics, resistance to many AMPs evolves slowly and is unrelated to resistance to other classes of commonly used antibiotics that are cross-resistant.
The difficulty of differentiating between true protein-coding sequences and false positives has been a technical limitation in the study of AMPs. Nonetheless, recent developments in metagenomic investigations have advanced significantly, utilizing machine learning methods to pinpoint smORFs encoding proteins that fall into particular functional groups. In recent work, metagenomic data of human gut microbiomes were used to identify about 2,000 AMPs using projected smORFs. There is great promise for finding AMPs from prokaryotes across various habitats, even though the human gut makes up only a small portion of the total microbial diversity.
Understanding AMPSphere
In this study, AMPs from the global microbiome, as reflected in public databases, were predicted and cataloged using machine learning. Numerous AMP diversity patterns were discovered through the analysis of 63,410 metagenomes and 87,920 high-quality microbial genomes. AMPs unique to various habitats rather than core genes in the pangenome were discovered as a result, culminating in the development of the AMPSphere, a collection of 863,498 non-redundant peptide sequences.
The ability to recognize functional AMPs from the global microbiome has been established by AMPSphere, a peptide synthesized from the global microbiome. Sequences cloaked in protein sequences, or c_AMPs, were synthesized and demonstrated antibacterial efficacy against clinically important ESKAPEE infections. These peptides were contrasted with encrypted peptides (EPs), which were computationally mined peptide sequences concealed within protein sequences. A preclinical animal model showed encouraging anti-infective effectiveness for the top contenders.
Limitations of the Study
The research focuses on peptides as a tool for comprehending the global microbiome. Peptides comprise up to 100 amino acids and are encoded by their genes. This global microbiome is represented in public databases; nevertheless, quality assessments are impacted by unequal coverage. The site will stay current with the release of new genomes and metagenomes. Error rates are higher when matching tiny sequences to huge databases, especially when mismatches occur. The usefulness of AMPSphere as a resource is demonstrated by results on the transmissibility of microbial strains and AMP density; nonetheless, complete validation is the main objective of future research. Peptides showed potential action against strains not examined in this study when tested against a range of bacteria both in vitro and in vivo.
Conclusion
Nearly a million potential AMPs, crucial for transcription and translation, are found in the human gut and the rest of the world’s microbiome. These AMPs were discovered by a machine learning study utilizing 87,920 high-quality microbial genomes from the ProGenomes2 collection and 63,410 publicly accessible metagenomes. As a result, AMPSphere was developed, which is an open-access database that contains 649 high-quality AMP families and 863,498 non-redundant peptides from 72 habitats, including the human gut, soil, and marine environments.
Article Source: Reference Paper | AMPSphere is available as a public online resource at https://ampsphere.big-data-biology.org/
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.