In a recently published paper by Peking University, China, researchers have outlined an analogy between the advancement of Artificial Intelligence (AI) technologies and the possibility of progressing Molecular Simulation (MS) techniques, which greatly aid scientists across diverse fields from physical sciences to life sciences. Moreover, the study articulates multiple aspects regarding the coherence of AI and MS, the consolidation of MS and AI, facets of AI-leveraged MS, and beyond.
Impediments in the Advancement of Molecular Simulation Techniques
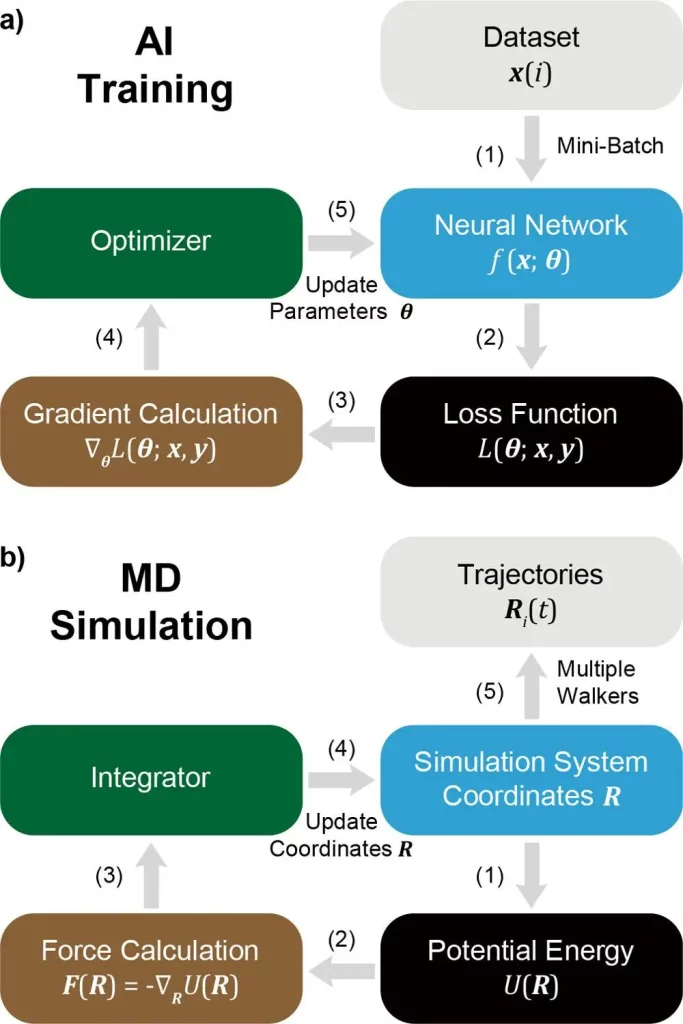
Image Source: https://doi.org/10.1021/acs.jctc.3c00214
Thermodynamics, dynamics, trajectories, steric and spatial characteristics of molecules in a real-life system are computationally emulated at the atomic level and visualized by typically computing laws and equations of physics in Molecular Simulation (MS), especially Molecular Dynamics (MD), which simulates movements of atoms illustrating the microscopic world, is a powerful and effective technique harnessed by scientists in material sciences, chemistry, physics, technology, biophysics, and bioinformatics, notably during Drug discovery and development.
First, the authors put forward the shortcomings of widely operated Molecular Dynamics (MD) software packages that, although they deliver reliable, high quality, effective and fast outputs, are still confined to accomplishing full-fledged enhancement owing to the programming language of the codebase.
The simulation packages consist of large specialized codebases written in C/C++ or FORTRAN and also include a significant extent of code duplication and hardcoded gradients, rendering it challenging to keep up with contemporary advancements in computational technology and infrastructure and integrate with optimized packages.
For instance, simulation software programs that are incompatible with GPUs will be bound to be outdated due to their inability to synchronize with the latest technologies. Also, in the generation of massive-scale parallel computing, the codes of Molecular Simulation programs are required to be substantially modified to sustain supercomputing clusters.
Whereas Artificial Intelligence has exhibited a revolution and surged towards solving complex problems like modeling protein structure and folding by incorporating breakthroughs like deep learning, most MD tools are having challenges accommodating the recent innovations. The paper emphasizes and acknowledges these instances and underlines the urgency of all scientific domains to embrace revolutionizing software and hardware infrastructures and AI methodologies for advancing Molecular Simulation tools.
Following the Trends of Artificial Intelligence and Computational Infrastructure Advancement for Upgrading Molecular Simulation Techniques
The researchers excellently point out the resemblance in the hindrances faced by AI in its early phase and MD currently, including cumbersome parallelization,hand-coded derivatives, incompatibility with state-of-the-art hardware, and deficient support for user-plugin functions. Although AI developers tackled these challenges amazingly, regardless of advanced MD Packages like AMBER or OpenMM, the problems are still largely unsolved in upgrading simulation strategies.
Likewise, most MS packages have significant amounts of software-specific codes written in various programming languages, causing difficulties in transferring functionalities from one package to another, and many functions or modules are duplicated across MS programs, making it strenuous to update. This state of affairs is reminiscent of the inception phase of machine learning when methodologies like automatic differentiation (AD) and automatic parallelization (AP) were not yet popularized. No wonder the AI community has supplied elegant solutions to such issues and made us witness a massive transformation in the virtual world.
Correspondingly, Automatic Differentiation (AD), which yielded rapid upgradation of Machine learning, can be utilized by current machine learning frameworks for determining atomic forces. Therefore, AD can also be implemented for calculating forces and force fields for MS. This would significantly reduce the complicated effort to calculate energy functions and the extensive computation required to integrate them into the MS codebase. For this drawback, due to the variety in energy functions and forms of hardcoded forces, MS programs have been separated from simulating Molecular Docking, Molecular Dynamics, protein structure prediction, and virtual screening.
Furthermore, a coherence between MD and AI Training is mentioned, pinpointing that MD can be treated as a subset or special case of AI. Here, for example, researchers describe the analogous nature of Particle coordinates in MD with Neural Network parameters, potential energy in MD with loss function in AI, and the equivalence of the MD integrator to the AI optimizer.
Regarding parallel computing, an automatic parallelization (AP) technique was developed for deep learning models, enhancing ease in programming and handling large-scale systems. The researchers enunciate that an AP-enabled MS platform would carry out multiple trajectories of simulations simultaneously on a single GPU or NPU, which is impossible for most conventional MD simulation software due to the coding complexity. An overall improvement in compatibility and extensibility of the MS package is essential for synchronizing with recent infrastructure development and exploring the full potential of MS in novel discoveries.
Conclusion
The exemplification of MindSPONGE in different parts of the paper illustrates the room for advancement of simulation strategies leveraging newer technology since the integration of AI and MS is in its infancy. The AI-enhanced simulation package MindSPONGE is based on a modern AI platform MindSpore comprises a modular architecture and Python API allowing differentiable programming, high-throughput computing, and automatic hardware transferability. For instance, the same research group formulated a parametric learning approach, IDM (Information Distilling of Metastability). It is integrated with MindSPONGE. IDM executes clustering, reduces the dimensionality of molecular systems, and can be implemented to comprehend mechanisms of protein folding.
Moreover, the researchers anticipate that with AI-enhanced MS, numerous drawbacks and constraints in simulating situations of chemistry and biophysics can potentially also be resolved by encompassing amassed data and physics rules. Throughout this study, the researchers wonderfully described the resemblance between AI training and MS and how MS packages can be advanced by taking inspiration from the innovations that boosted AI technology and encouraging the different scientific communities to adopt and democratize the technological revolution by updating MS packages.
Article Source: Reference Paper
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.