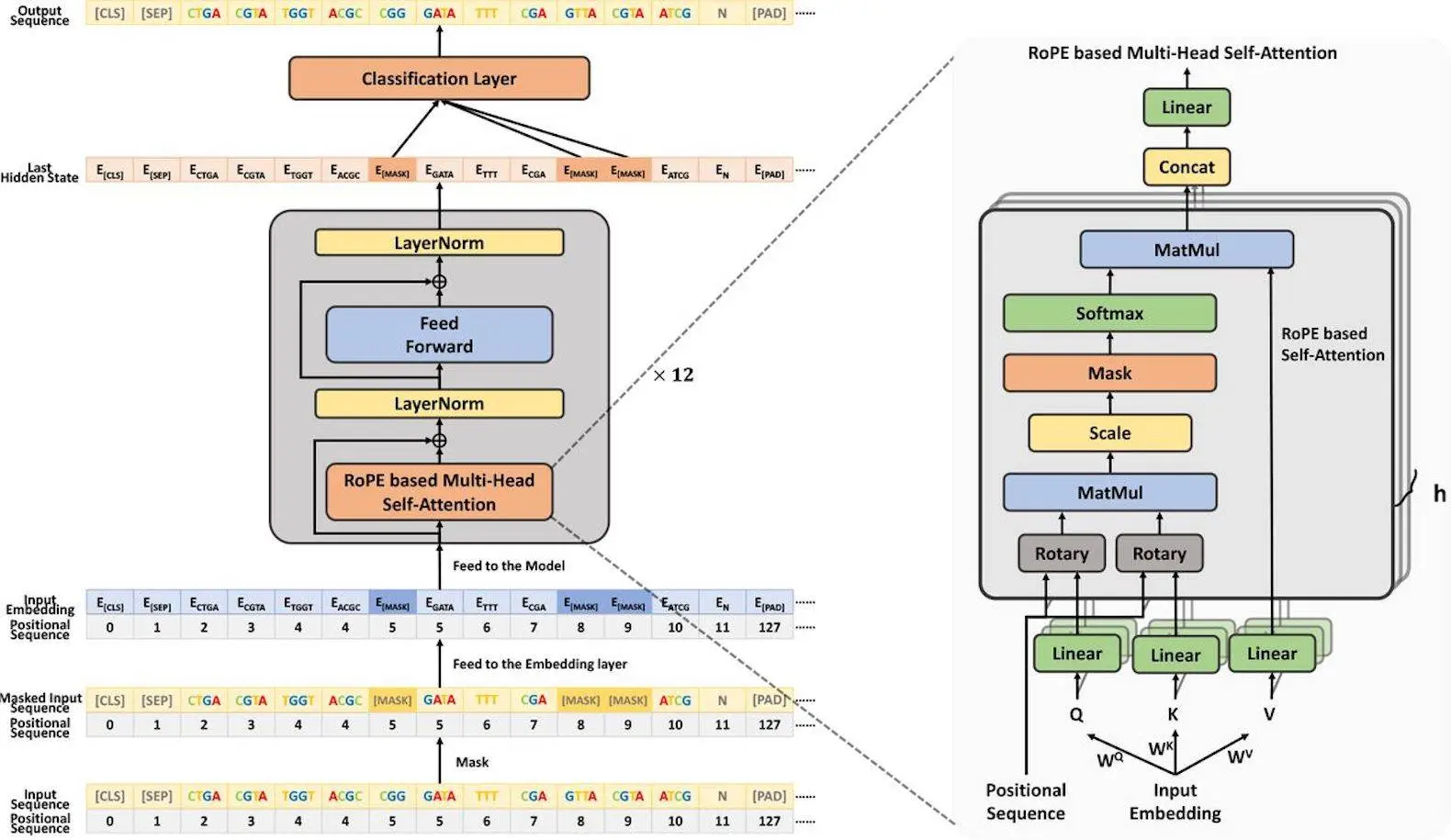
Faculties of the Mathematics Department at the University of Warsaw, Poland, have presented a review of the past two years progress of Artificial Intelligence (AI)-mediated approaches to boost Antimicrobial Peptide (AMP) designing-related research works, which are absolutely necessary to overcome the global health hazard manifested by the upsurge of antibiotic-resistant pathogens. The review article offers comprehensive summaries regarding different methods strategized by AI in AMP design, characterization of the tasks AI can execute to drive AMP research, different approaches to evaluation of AMP discovery, and also inscribes valuable insights for further improvement.
Encountering Antibiotic Resistance: A Pressing Need
Microbial resilience against antibiotics is becoming more prominent and severe day by day. With antibiotics, bacterial infections were no big deal to encounter, as they were supposed to be a century back. Antibiotics are prescribed and capitalized on for their magical curing capacity. Antibiotic abuse for decades has led to this rapid surge of antibiotic resistance among infective organisms and is a major threat to the healthcare system. Pathogens have gradually developed strategies to escape the effects of antibiotics and are emerging as too notorious to be controlled.
Finding immediate alternatives to tackle microbial infection and antibiotic resistance is a burning requirement for this decade; otherwise, scientists expect much more havoc than today’s. Many laboratories across the globe have been in search of safer substitutions for years. One of the most harnessed solutions utilized and commercialized nowadays is multidrug therapy, where a combination of antibiotics is prescribed. Sadly, the report of Multi Drug Resistance (MDR) Bacteria is not sparse but rather a very common occurrence.
Antimicrobial Peptides as an Alternative to Encountering Antimicrobial Resistance
Newer research in fighting this health emergency has shown positive results, but most of them aren’t inexpensive and, importantly, can’t be democratized like antibiotics. In this aspect, Antimicrobial Peptides have illuminated rays of hope as an alternative pharmaceutical for their promising characteristics in combating drug-resistant pathogens. Antimicrobial peptides are small bioactive peptides that mostly have a net positive charge and possess less than 100 amino acids. Although AMP’s success rate in clinical scenarios is limited, its prospects encourage scientists to design more effective AMPs.
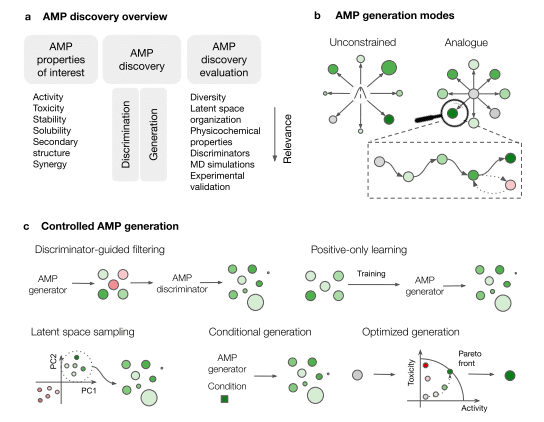
The clinical potential of an AMP is judged by evaluating its activity, toxicity, stability, and synergy, which are expressed as the Fractional Inhibition Concentration (FIC) index. One commonly used protocol to detect antimicrobial activity is the Minimum Inhibitory Concentration (MIC) or Minimal Bacterial Concentration (MBC) assay, which is also done for the AMPs to check their activity rate against bacterial strains. Peptide toxicity is typically measured by assaying hemolytic activity and cytotoxicity, which is performed against different cell types. HC50, a hemolysis test, determines the least peptide amount that can cause disruption of 50% of human erythrocytes. Moreover, numerous databases reposit the activity and toxicity measurements, along with sequence and structure.
Insights on AI-Driven AMP Research: Approaches, Evaluation Strategies, and Limitations
Notably, the study provides an essence of AI technological development focusing on AMP design in the last two years.
- Antimicrobial Peptides are mostly represented by their sequences of amino acid codes. Apart from that, properties like amino acid composition, physicochemical attributes, sequence similarity, and structural details like secondary structure, molecular fingerprints, and atom-type connectivity are also noticed to be used while fabricating models. Pretrained language model-derived sequence embeddings have recently proven effective for AMP discovery.
- AI-driven AMP discovery methods mostly operate in a supervised setting, with positive and negative datasets used for training. In both cases, there can be possible biases, such as in the former cases where the whole collection of peptides from AMP databases, regardless of their target species or strain, are taken into consideration; most of the AMPs are tested on E. Coli strains only. In negative datasets sampled from UniProt, excluding entries matching keywords such as antimicrobial, antibiotic, secreted, and similar often renders sampling method-related biases. While the models take into account aspects like activity or toxicity, they often encounter problems regarding measurement units.
- Most importantly, the paper discusses two groups of AI methods for AMP discovery: discrimination and generation. AMP discriminators prevalently classify peptides broadly as either AMP or non-AMP; for instance, AMPlify, AMP-BERT, VGG16-AMP, etc. The authors enunciate that multi-label classification models often face issues due to limited training data. MIC prediction methods have shown better outcomes in this regard. Some other AMP discriminator strategy models consider genomic information of the strain, FIC, hemolytic or cytotoxicity, solubility, activity, etc.
- Generative AI models are also leveraged largely for AMP discovery. In the unconstrained generation mode, peptides are freely sampled from the model de novo implementing GAN, Recurrent Neural Network (RNN), and Graph Neural Network (GNN), as well as all VAE models. The analog generation task is executed solely using VAEs. The controlled AMP generation process essentially considers controlled properties of peptides like activity, toxicity, microbial target, target mechanism, hydrophobicity, secondary structure, as well as sequence length and is obtained by approaches including discriminator-guided filtering, positive learning, latent space sampling, conditional generation, and optimized generation.
- Commonly, the discriminative model uses evaluation metrics such as the area under the curve (AUROC) for classifiers or the root mean squared error (RMSE) for regressors. Generative models are evaluated by applying discriminators to the generated peptide sequences for assaying the properties of generated AMPs. The authors point out that the limited accuracy of the existing discriminators may be biased or provide an exaggeratedly optimistic evaluation.
Conclusion
The paper provides insightful overviews of the domain that needs to be emphasized for future research in the field of novel AMP design with pharmaceutical benefits. The criticism of the gap in validating the activity of AI-designed AMPs in the experimental scenario is significant of all. More focus should be on the in-depth characterization of prospective peptides in the wet labs. Besides, other intuitive recommendations provided by the researchers include the formulation of community-wide accepted benchmarking datasets combined with open sharing of code, consolidating every possible data regarding AMP to generate novel models with larger datasets, emphasizing explicit modeling or correcting for the data noise and scarcity; clustering the intrinsic similarities of AMPs; ranking the generated peptides, exploiting Molecular Simulations for evaluation; devising optimization strategies followed by generation. Thus, there is still multifarious scope for improving and accelerating AMP research that can help to overcome antimicrobial resistance-associated health risks.
Article Source: Reference Paper
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Learn More:
Aditi is a consulting scientific writing intern at CBIRT, specializing in explaining interdisciplinary and intricate topics. As a student pursuing an Integrated PG in Biotechnology, she is driven by a deep passion for experiencing multidisciplinary research fields. Aditi is particularly fond of the dynamism, potential, and integrative facets of her major. Through her articles, she aspires to decipher and articulate current studies and innovations in the Bioinformatics domain, aiming to captivate the minds and hearts of readers with her insightful perspectives.