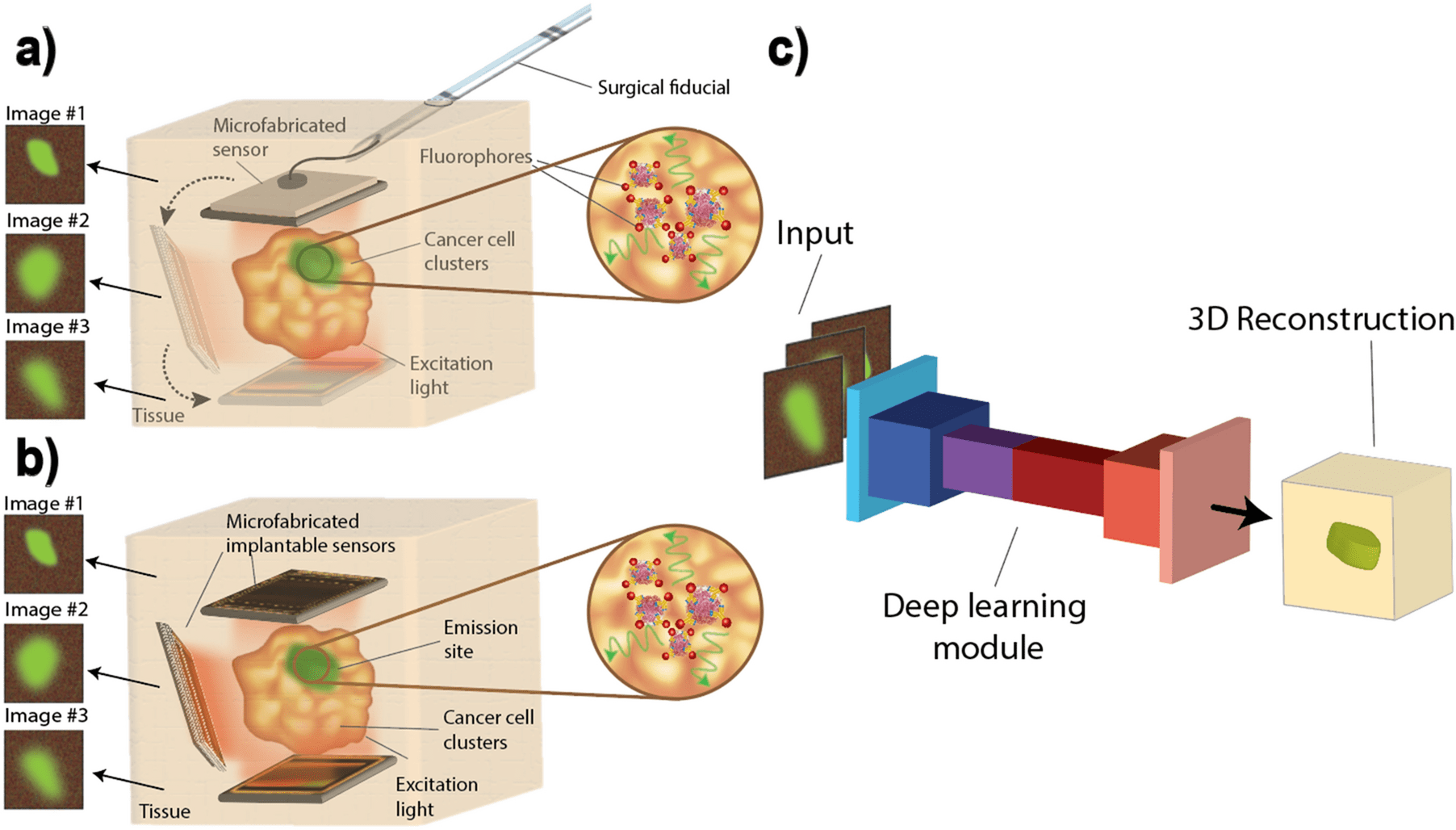
Microscopic imagers may be used in oncology to track and observe cell motions and dynamics, indicating tissues' real-time response to therapy.
Scientists from the Department of Electrical Engineering and Computer Sciences, University of California, Berkeley, reconstruct cellular images in 3D from microfabricated imagers using fully-adaptive deep neural networks. This research is the first work that consolidates deep neural networks for depth estimation from singular cellular level images.
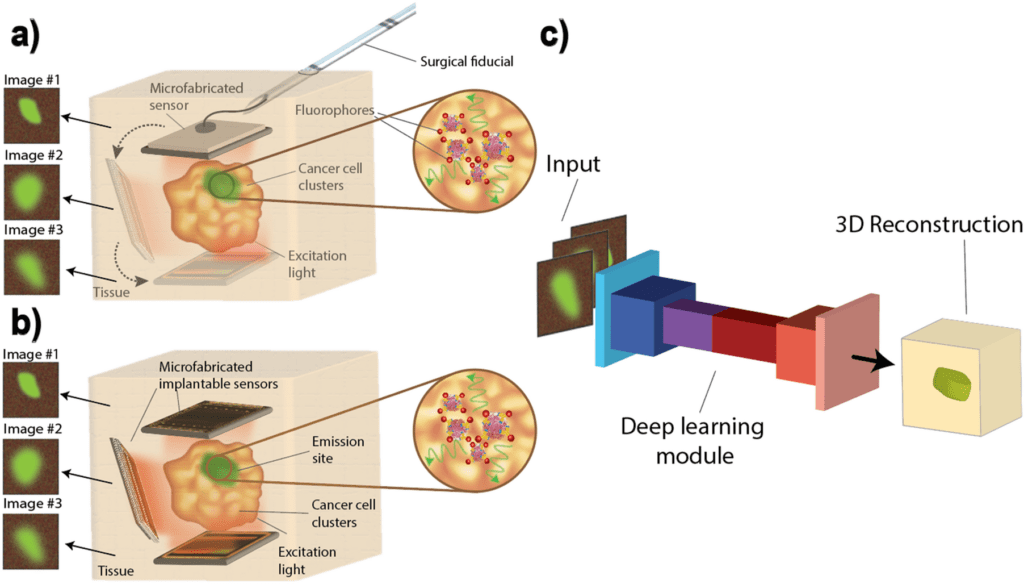
Image Source: 3D Reconstruction of cellular images from microfabricated imagers using fully-adaptive deep neural networks.
Millimeter-scale, multi-cellular level imagers empower varied applications, ranging from intraoperative surgical navigation to implantable sensors.
However, the tradeoffs for miniaturization compromise resolution, making extricating 3D cell locations challenging — vital for tumor margin assessment and therapy monitoring. This work explains three machine learning-based modules that extract spatial data from single image acquisitions by the utilization of custom-made millimeter-scale imagers.
The neural networks were trained on synthetically generated (by the utilization of Perlin noise) cellular images. The first network was a convolutional neural network estimating the depth of a single layer of cells, the second was a deblurring module rectifying the point spread function (PSF).
The last module extracted spatial data from a single image acquisition of a 3D specimen and reconstructed cross-sections by giving a layered “map” of cell locations. The maximum depth error of the first module was found to be 100 µm, with 87% test accuracy.
The PSF correction of the second module accomplishes a least-square error of just 4%. The third module generated a binary “cell” or “no cell” per-pixel labeling with an accuracy range going from 89% to 85%.
This work exhibited the synergy between ultra-small silicon-based imagers that empower in vivo imaging but face a compromise in spatial resolution and the processing power of neural networks to accomplish enhancements past conventional linear optimization techniques.
The Preference for a Less Complex Imaging Platform with a Smaller Form Factor
Visualizing the 3D location of the fluorescently labeled tumor cells in vivo is vital for intraoperative navigation to recognize tumors underneath the tissue surface and harbored in deeper sites like lymph nodes.
Ordinarily, this process is done post-operation in a laboratory setting by the utilization of targeted fluorescent probes and markers to recognize any disease from the tumor bed sample. This cycle is incredibly tedious and requires a few days to return results, potentially risking the result of the treatment.
Latest engineering advancements have brought about a few novel imaging platforms that permit this cycle intraoperatively and concurrent with the surgery.
Be that as it may, these instruments are essentially cumbersome and not practical with the present negligibly invasive surgeries, particularly in complex and difficult-to-access tumor cavities.
While these huge instruments depend on sizable optics and lenses for their high resolution and unwavering quality, they can’t be miniaturized and made practical for surgical settings because of the rigid optical equipment.
Furthermore, such massive instruments naturally do not allow for evaluation of treatment response, where cell migration into tissues is vital to be monitored progressively in real-time and outside surgical settings — another fundamental application.
Along these lines, a considerably less complicated imaging platform with a smaller form factor is preferred. For example, the latest advancements in microscopy and light-field microscopy have empowered imaging of smaller features and imaging inside the tissue.
Notwithstanding, these strategies require specialized optical equipment incompatible with a negligibly invasive procedure.
Miniaturization of these platforms into electronic micro-imagers empowers their placement in difficult-to-access regions, opening the capability not only to visualize microscopic disease intraoperatively in cavities up to a few millimeters deep, yet in addition to monitor cell dynamics and evaluate treatment progressively in real-time and in vivo, with a network of wirelessly powered implants.
A Requirement for Enhanced, Custom-made Optical Fibers, and Lenses
While the ultra-small imagers are not difficult to integrate into surgical environments, unlike their bigger counterparts, they miss the mark on sufficiently high resolution.
Decreasing the form factor of the imager forces a stringent limit on the size of the optical filters and focussing lenses being utilized, limiting their performance and the image resolution.
Smaller imagers required for in vivo utilization frequently have a higher background level and a lower focusing ability. Safety limits likewise limit the total photon budget permitted inside the system, further constraining the abilities of fluorescence microscopy in vivo.
To acquire dependable 3D data by the utilization of imaging instruments with these small form factors, enhanced custom-made optical filters and lenses are required to replicate similar performance as their bigger counterparts.
Nonetheless, these are often difficult to manufacture, and the image quality remains suboptimal compared to bench-top microscopes. In this manner, computational methods that can enhance images from small form factor devices are in need.
The Point Spread Function or PSF
Conventional image processing strategies involve variations of deconvolution, surface projection algorithms, and noise enhancement methods, and the core of this large number of techniques depends on a linear transformation of the image that doesn’t rely upon anything besides the raw image data and the point spread function (PSF) of the imaging device and is procedurally heedless to any prior knowledge of the specimen being observed.
PThe PSF, otherwise called the transfer function of the imaging system in the spatial domain, depicts the response of an imaging system to a point source of illumination. In any linear image formation process, for example, fluorescence microscopy, the final image is a linear superposition of a series of point sources convolved with the PSF.
Therefore, the original object can be retrieved by deconvolving the image with the PSF of the imager.
Drawbacks of the Deconvolution Method and Circumventing Them
One of the downsides of the deconvolution method is the calibration required for the derivation of parameters of the deconvolution function for images taken from every depth, which limits image processing speed.
This technique presents challenges for recovering images consisting of overlaid cell foci from various depths since each parameter requires optimization for a certain imaging depth.
Additionally, the PSF, as a low pass transfer function, eliminates high-frequency components of the original image, thus bringing about a loss of sharpness and obstructing the generation of a completely recovered image after deconvolution.
In any event, for situations where the image is mainly attenuated by the PSF response and still retains the greater part of the high-frequency components, applying inverse PSF amplifies high-frequency noise degrading the recovery of the original image.
Similar to deconvolution, every other linear image processing method would experience the ill effects of similar issues.
A more agile methodology is required to circumvent these limits, particularly a non-linear post-acquisition processing module that could consolidate the physiological and spatial data of similar tissue specimens inside itself.
The module can leverage this additional information to restore the sharpness and resolution of the micro-imager’s suboptimal images and give insight into the 3D position of cells. Of every accessible architecture, deep learning modules can be by a wide margin the most proficient at capturing physiological information of cellular images.
Applications for Cancer Imaging Using Deep Learning
Deep learning consolidates multiple layers of non-linear transformations, superposed with a complex yet structured network of coefficients to make powerful processing modules that perform exceptionally complex tasks, like image enhancement, image classification, and feature extraction.
Deep learning permits breaking the tradeoffs of fluorescence microscopy and utilizing the computational models to augment hardware complexity and improve optical limits by using an enormous training data collection to build the network.
By the utilization of adaptive network architectures like residual neural networks (ResNets) and convolutional neural networks (CNNs), the scientists present a few applications for cancer imaging using deep learning to enhance the resolution and ability of custom-made micro-imagers.
Challenges in Using Neural Networks in 3D Cellular Imaging
A vital test to involve a neural network in 3D cell imaging of tissue is compiling a training dataset since training deep neural networks requires access to an enormous set of training data from diseased tissue for every particular application.
The impracticality of acquiring an enormous dataset, taken at different depths, from tissue motivates the synthesis of a diverse dataset of cancer cell images in view of the morphology of real-life tissue samples to leverage prior knowledge of the cancer cells.
The synthesis technique should be parameterizable to permit the generation of an arbitrary large dataset by random selections of parameters that lead to images that are accurate representations of actual cells.
Addressing the Challenges
To address the challenges in the utilization of neural networks in 3D cellular imaging, the scientists first present a technique to generate an enormous training set mimicking real-life specimens, subsequently permitting their deep neural networks to be trained.
To replicate the 3D structure of the tumor, stacks of various layers of cells with 250 µm spacing within 1 mm from the sensor were created. Since the lensless custom imager was designed for contact imaging of tumor margins, 1 mm was set as the limit to show evidence of the concept.
Then, the researchers present three modules entrusted with recognizing the depth of cell clusters – measured as the distance of the sample from the imager-deblurring and improving the sharpness of the image, and lastly, identifying cell presence inside each layer of the specimen in 3D stacks.
The last module incorporated a novel method for imaging utilizing not one but two sensors viewing tissue from various points to consider three-dimensional imaging of the specimen and giving insights into the spatial distribution of the cells in the sample.
The Endpoint
Getting high-resolution, cellular level data from in vivo images of tissue is vital in oncological applications. This research is the first work that consolidates deep neural networks for depth estimation from singular cellular level images. A large synthetic dataset representative of genuine cancer cell images empowered training of deep neural networks for single and multilayer depth estimation and image deblurring and resolution improvement.
The accuracies of 87% for single-layer depth estimation, 95.8% for deblurring, 93.8%, and 86.5% for cell localization and depth estimation of non-overlapping and overlapping stacks of various layers of cells were accomplished, respectively. The novel imaging platform introduced here leveraged placing two sensors in tissue empowering high depth estimation accuracy for intraoperative applications, which was made entirely possible by the ultra-small form factor of the custom-designed microchip sensor.
Article Source: Najafiaghdam, H., Rabbani, R., Gharia, A. et al. 3D Reconstruction of cellular images from microfabricated imagers using fully-adaptive deep neural networks. Sci Rep 12, 7229 (2022). https://doi.org/10.1038/s41598-022-10886-6
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.