Researchers from Tartu University have put forth a semi-supervised machine learning framework to detect invasive ductal carcinoma using a small amount of labeled breast cancer histological data.
Breast cancer is one of the most common cancers. According to the world health organization (WHO), nearly 2.3 million women worldwide have been affected with breast cancer, with 685,000 deaths in the year 2020. Breast cancer exhibits complex histological and molecular heterogeneity. Invasive Ductal Carcinoma (IDC) occurs when the abnormal cells growing in the lining cells (epithelium) invade the breast tissues present beyond the duct walls. IDC is the most common breast cancer type, contributing to 80% of the total breast cancer cases. Lumps in the breast and other morphological features such as tumor size and shape play a critical role in breast cancer diagnosis.
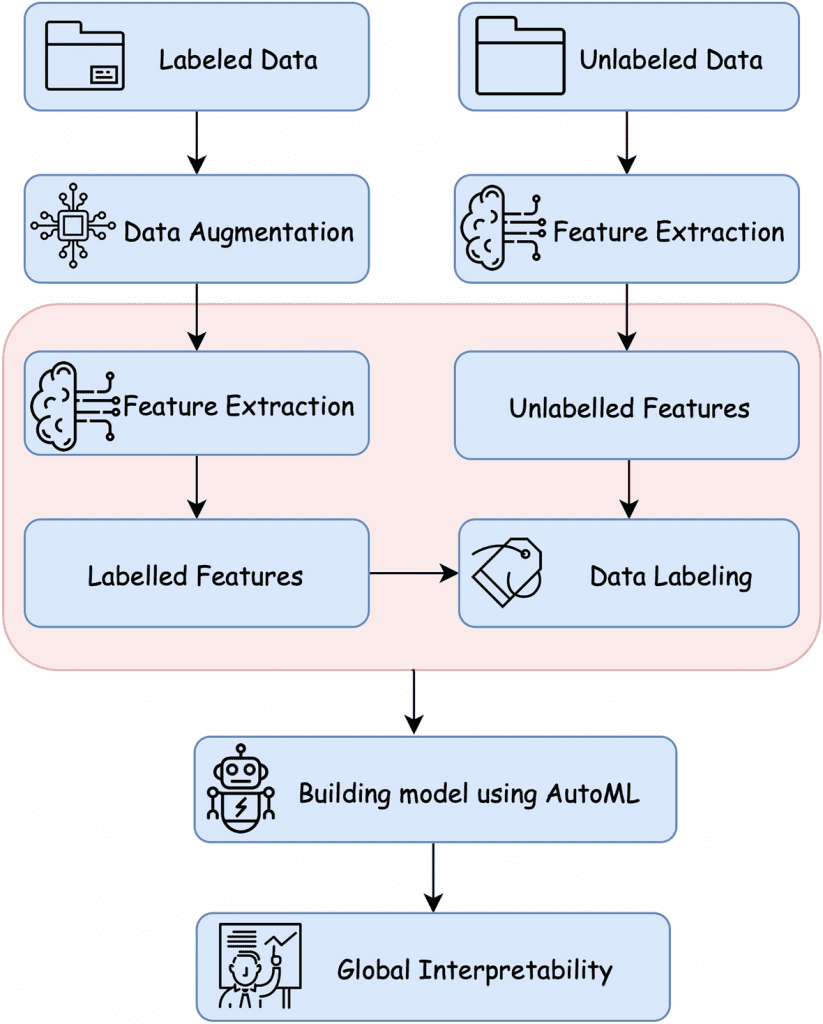
Image Source: https://doi.org/10.1038/s41598-022-20268-7
Medical diagnoses have taken up deep learning approaches for image recognition and classification. Machine learning framework approaches such as TensorFlow and PyTorch have eased the ability to develop deep learning models, but with the disadvantage of needing high-quality labeled data with accurate parameters and a structured architecture.
Generative Adversarial Networks (GANs) are a deep learning-based model that has gained significant traction in the medical domain for its powerful image-generating tasks. But due to the lack of labeled medical image data available, semi-supervised learning approaches are preferred to traditional supervised learning. Semi-supervised learning is a machine learning-based approach that combines a small amount of labeled data with a large amount of unlabeled data. Self-labeling of such data is achieved by techniques such as self-training and co-training. The co-training algorithm is more prevalent where each sample is detailed based on two different feature views that provide complementary information on the same sample. Samples with the most confident predictions on the unlabeled data are then added to the labeled data. Machine learning results can be interpreted using global and local techniques. The global technique focuses on comprehending the model on a global scale, whereas local predictions are interpreted in a single sample.
With these approaches in mind, the current study has hypothesized a five-stage global semi-supervised learning approach for detecting breast carcinoma to combat the low availability of labeled data. The five major stages include data augmentation, feature selection, splitting, and co-training data labeling, neural network modeling, and the interpretation of neural network prediction. A small amount of labeled data was collected from digitized breast cancer (BCa) histological slides from 162 women diagnosed with invasive ductal carcinoma at the Cancer Institute New Jersey and the University of Pennsylvania Hospital.
Findings
Current techniques to identify abnormalities in medical data use manually annotated images for training the framework. But in the El Shawi et al. study, a neural network was trained using labeled data to label the unlabeled data generated by a Deep Convolutional Generative Adversarial Network (DCGAN) for IDC detection. The trained neural network model has achieved a balanced accuracy of 0.865 AUC and an accuracy of 85.75%, which exceeds the balanced accuracy of 0.843 AUC and an accuracy of 81.28% when using only a control, Dtrain. For higher performance and better labeling, techniques such as Principal Component Analysis (PCA), Linear Discriminant Analysis (LDA), and VGG-16 were employed, with VGG-16 showing robust results of 0.815 AUC.
Results have also shown that increasing the unlabelled data size significantly affected the model’s performance, where splitting the labeled dataset into extremely small instances is critical to labeling and is more sensitive to noise. An AutoML framework was employed to choose the best possible architecture and accurate parameters for the datasets used in the study. The suggested architecture outperforms the state-of-the-art model by Cruz-Roa et al. (balanced accuracy of 0.87 and F-measure of 79%) and Janowzyk et al. (balanced accuracy of 0.85 and F-measure of 76%). The current study has only used 10% of the labeled data yet has provided significant results compared to other study designs.
The computational model classified malignancy from normal by the contrast in nuclei and by comparison with breast tissue patches. This also goes with the NHG grading system (The Nottingham Grading System, a prognostic factor in breast cancer widely used for clinical diagnosis), where hyperchromatism is a sign of breast cancer symptoms for diagnosis. The technique is also locally adaptable and is able to provide predictions on breast tissue patches. Local predictions are made by determining the closest patch and then calculating the BR scores (Br score is similar to Pearsons correlation with the scores ranging from 1 to -1); the scores were found to be in alignment with the NHG system as well further delivering credibility to the hypothesized approach.
Final Thoughts
The proposed model provides a novel semi-supervised deep learning approach to detect invasive ductal carcinoma where the model utilizes small amounts of labeled data for training and an abundance of unlabeled data. The results showed that the performance of the neural network model improved when the unlabeled data was combined with the original labeled ones. This study overthrows the limitation of the scarcity of labeled data in the medical domain.
The synthetic images generated by DCGAN and unlabeled data in training increased the overall performance. To develop clinicians’ trust in the deep learning model, the global technique of nuclear segmentation was interpreted and is in line with the diagnosis hallmarks.
One of the significant limitations of the study is that querying a particular set can cause biased interpretation while failing to address the right set of concepts. Other drawbacks include an unlimited number of queries, which leads to an indefinite number of meaningful variations, making it hard to provide many examples to develop trust in these concepts. Authors have also suggested a possible extension of their work to extract needed information from the provided data, thus easing histopathologists and medical diagnoses.
Article Source: Reference Paper | Data Availability: IDC Batch-based Data | Nuclear Segmentation Data | Code Availability
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Shwetha is a consulting scientific content writing intern at CBIRT. She has completed her Master’s in biotechnology at the Indian Institute of Technology, Hyderabad, with nearly two years of research experience in cellular biology and cell signaling. She is passionate about science communication, cancer biology, and everything that strikes her curiosity!







