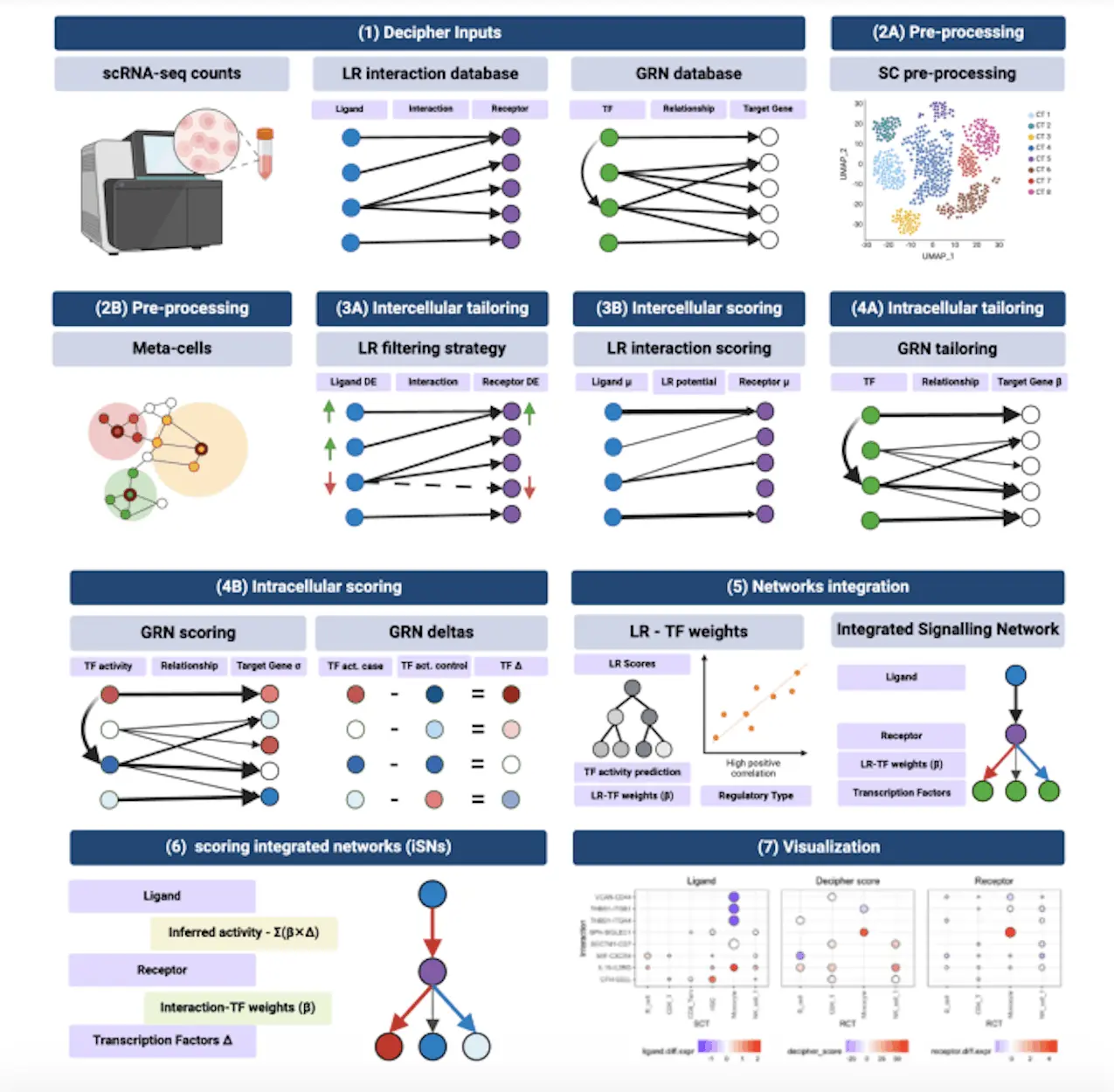
Scientists from the University of California, Berkeley have developed a machine learning-based model that predicts protein fitness from the sequence.
Protein fitness is a broad term that includes a wide range of protein properties, such as stability, enzyme activity, and ligand binding. Models of protein fitness based on machine learning typically learn from either unlabeled, evolutionarily related sequences or variant sequences with experimentally measured labels. The researchers propose a simple combination approach that outperforms more sophisticated methods. This method combines ridge regression on site-specific amino acid features with one probability density feature from evolutionary data modeling. Overall, the variational autoencoder-based probability density model performed the best.
Proteins naturally serve many vital jobs supporting life, but they have also been co-opted for human pursuits such as genome editing. Laboratory-based directed evolution and computational, physics-based rational design are the two most common approaches to protein engineering. In silico screening can be supplemented by machine learning-based methods that predict protein fitness from the sequence. The emphasis in this research is solely on machine learning methods for protein fitness prediction.
There have been two main machine learning-based methodologies for estimating protein fitness models. The first approach is to use evolutionary data. Such methods begin with a query protein with the desired feature and search through databases of naturally occurring proteins to find a collection of related proteins connected mainly by sequence homology and are presumed to be enriched for the same attribute as the query sequence. Then, a probability density model is estimated; finally, sequence density evaluations are utilized to forecast the relative fitness of protein variants of interest.
In the second major machine learning technique, supervised regression models are trained using variation sequences and fitness labels determined in the lab. A supervised data set may contain hundreds to hundreds of thousands of examples, depending on the protein and the attribute of interest. The laboratory assay could be a relatively direct evaluation of the fitness of interest or a crude proxy, with the collection of variant sequences limited to one or two mutations away from a query sequence. Only a few sequence locations in the labeled variant sequences may differ. These data sets are often not sufficiently rich to define protein fitness landscapes because of their limited construction, but they provide a solid starting point.
Several proposals have recently been made to integrate these two machine learning strategies: weak-positive only learning on evolutionarily related sequences and supervised learning on assay-labeled variant sequences. The researchers refer to this configuration as weak-positive semi-supervised learning. Property labels can only be evaluated for hundreds of protein sequences in many instances of practical importance. It is critical to merge both data sources, especially in such a regime (but not only in this one). This straightforward method can be used for any evolutionary probability density model and adds little computational effort.
A total of 13 published machine learning algorithms for protein fitness prediction that use evolutionary data, assay-labeled data, or both were evaluated in the study. And the Evolutionary Scale Modeling (ESM)-1b model was chosen, pre-trained on UniRef50 (unsupervised), and then fine-tuned using assay-labeled data (supervised) because it was proven to have better performance on several prediction tasks when compared to other transformers at the time of the assessment.
Beginning with a simple baseline approach, the researchers developed a method that uses evolutionary and assay-labeled data and is competitive with far more expensive and complex approaches. The researchers employed ridge regression on one-hot encoded site-specific amino acid characteristics enhanced with one extra feature—the sequence density evaluation—for any already-trained evolutionary probability density model. Given an already-trained probability density model, the augmentation comprises just training a linear regression model, incurring negligible computing load.
For each data collection, 20% of the labeled data available was used for test sets. In addition to these predetermined training data set sizes, the remaining 80% of the available data was used for a more direct comparison with TLmutation and ESM-1b4, which is known as an 80/20 train/test split. The researchers employed “five-fold cross-validation on the training data set for hyper-parameter selection when it was computationally viable and otherwise held out 20% of the training data as validation data.” The authors averaged performance over 20 random seeds that randomized the data divisions for each training data set size, including the 80/20 split.
In this research, two measures of predictive model performance were used: (1) a Spearman rank correlation between actual and predicted fitness values, and (2) a ranking measure from the information retrieval community called normalized discounted cumulative gain (NDCG), which gives high values when the top predicted results are enriched for truly high fitness proteins—that is when the model accurately predicts the ranking of the fittest sequences.
When existing methods were compared to each other, and the augmented Potts model, the latter model consistently outperformed existing methods, including more sophisticated methods such as eUniRep regression. The only exception was that the evolutionarily based Variational AutoEncoders (VAE) beat the augmented Potts model on the double-mutant data in the low data regime. When average performance is broken down into specific data sets, the augmented Potts model is competitive on most data sets.
Regardless of the density model augmented, the augmented model consistently beat the comparable non augmented existing technique, irrespective of the training data set size.
Sixteen of the nineteen supervised data sets contained only single mutants of wild-type, and the three data sets had higher-order mutants, namely the green fluorescent proteins (GFPs), Poly(A)-binding protein (PABP) RRM domain, and ubiquitination factor E4B (UBE4B), U-box domain data sets were then more closely studied. Further, the models were trained using single-mutant labeled data and then tested on single, double, triple, and quadruple mutants. The models were also trained on single-mutant and double-mutant data. The results were qualitatively comparable to training on only single-mutant data; however, the relative difference in performance between any two models generally decreased as more data was added. It was found that simply using the edit distance itself is predictive of fitness relative to the machine learning models examined in data sets where the assay-labeled variants encompass a variety of edit distances from the wild-type. In contrast to GFP, mutation count is not as significantly predictive of UBE4B U-box domain data, likely because the mutational impacts are more heterogeneous in the sense that they include more varied, nonadditive, or both effects.
The augmentation methodology readily incorporates structure-based features or predictions from existing approaches as extra regression inputs. To provide a feel of where such future work might go, FoldX-derived stability features were added to the models on the three data sets with higher-order mutations that increased performance on GFP relative to the augmented VAE, but not on PABP-RRM or UBE4B.
The preliminary findings of the research suggest that whether structure-based characteristics give additional information to evolutionary and assay-labeled data may depend on the property of interest and the quality of available structures. Thus, machine learning-based methods for protein fitness prediction that use both evolutionary and assay-labeled data were compared. A simple baseline approach was introduced, in which evolutionary density models are augmented with supervised data in a linear regression model on site-specific amino acids acid features.
One would be confused at first as to how linear models with only site-specific features can generalize to mutations not detected during training, let alone outperform nonlinear models in this job. The model learns about the relevance of each location through regularisation, despite the fact that each amino acid at every position has its parameter. Thus, if the effects of several mutations at the same site are in the same direction, regularised linear models can generalize reasonably well.
Researchers anticipate richer models to emerge when assay-labeled datasets get larger and more varied (i.e., encompassing more of protein space). Furthermore, distinct machine learning algorithms are likely to be required for smaller and larger labeled data regimes, both of which are anticipated to remain important in the future of protein engineering.
Story Source: Hsu, C., Nisonoff, H., Fannjiang, C., & Listgarten, J. (2022). Learning protein fitness models from evolutionary and assay-labeled data. Nature Biotechnology, 1-9.
Data availability – https://doi.org/10.6078/D1K71B
Code availability – https://github.com/chloechsu/combining-evolutionary-and-assay-labelled-data
Swati Yadav is a consulting intern at CBIRT. She is currently pursuing her Master's from Delhi Technological University (DTU), Delhi. She is a technology enthusiast and has a keen interest in the latest research on bioinformatics and biotechnology. She is passionate about exploring new advancements in biosciences and their real-life application.