Scientists from Ludwig Maximilians University Hospital Munich, Germany have developed a new machine-learning method for classifying hard to diagnose sinonasal tumors. The DNA methylation information was utilized to thoroughly examine the molecular patterns of sinonasal undifferentiated carcinomas (SNUCs), which were inaccurately defined and contested in earlier investigations.
Sinonasal tumors have a variety of histopathologies and originate from sinonasal cavity histologic elements. Tumors in the paranasal sinuses and nasal cavity cover a wide range of tumor forms, despite their small size. They are difficult to identify since they frequently don’t have any particular pattern or appearance. It’s especially true for cancers called sinonasal undifferentiated carcinomas (SNUC).
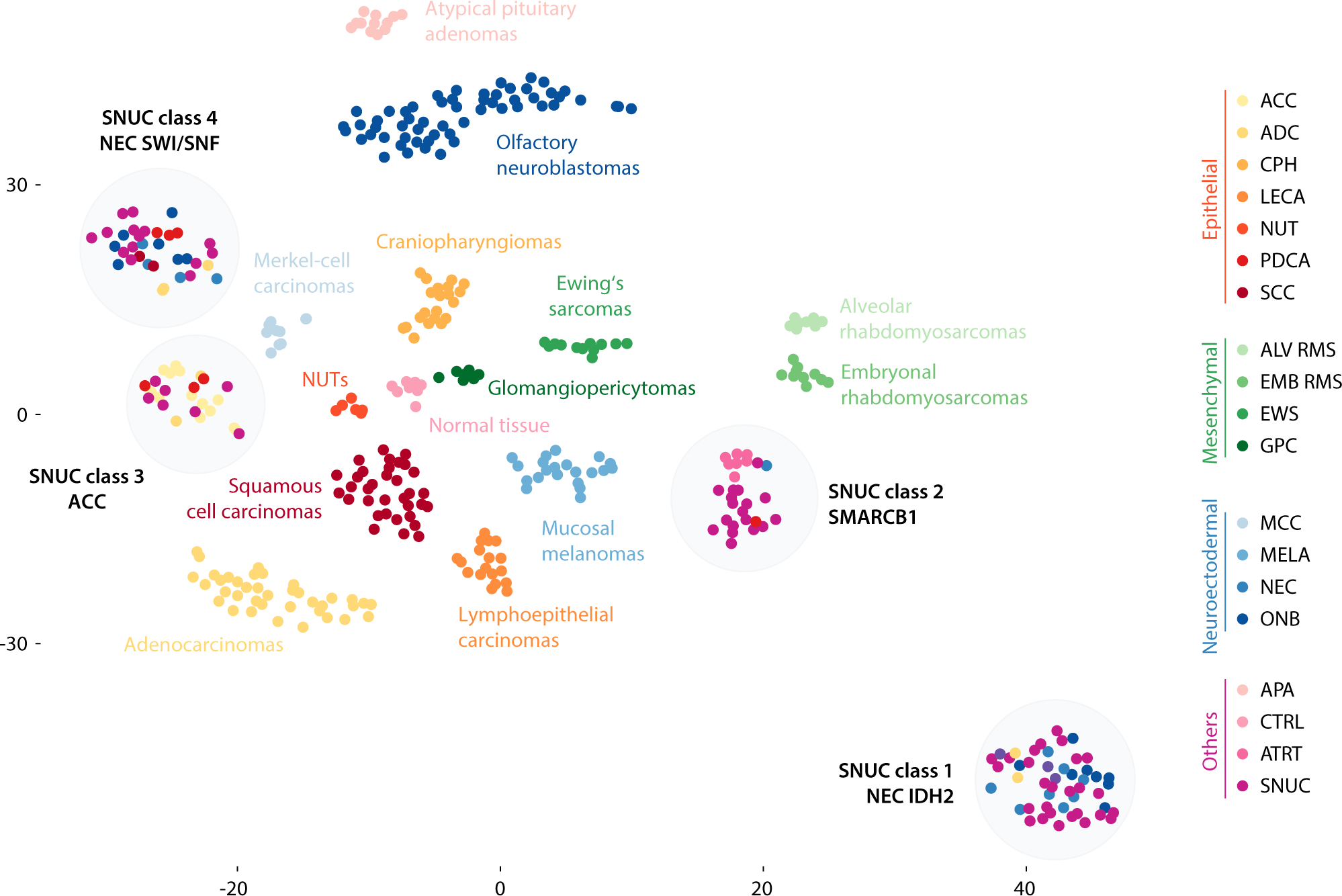
Furthermore, it is demonstrated that sinonasal tumors with SNUC morphology may be classified into four separate molecular groups based on their epigenetic, mutational, and proteomic profiles, not as undifferentiated as their present name would have us believe.
DNA methylation is a type of epigenetic alteration that controls how genes are expressed. It has been demonstrated that DNA methylation sequences are very tissue-specific, and it plays a crucial role in the differentiation of various cell types.
Gene activity is controlled in large part by chemical alterations to DNA. This involves the process of adding an additional methyl group to the DNA building blocks, known as DNA methylation.
Olfactory neuroblastomas and a cohort of sinonasal carcinomas had their DNA methylation profiles examined, and the results indicated that IDH2 mutant and SMARCB1 deficient carcinomas likely indicate epigenetically separate classes. Additionally, due to their epigenetic similarities, it has been proposed that IDH2-mutated neuroendocrine carcinomas and IDH2-mutated SNUCs may both refer to the same organism.
Classification of sinonasal tumors based on DNA methylation
It is intended to demonstrate, using an independent test set, that the DNA methylation-based classification system can accurately categorize samples with SNUC morphology without requiring further molecular analysis. In addition, the classifier accurately classified two samples that were previously identified as sinonasal squamous cell carcinomas as lymphoepithelial carcinoma and NUT midline carcinoma, correspondingly.
Sinonasal undifferentiated carcinoma (SNUC) class reclassification
The available histological and molecular data on tumor specimens were classified into the four SNUC classes identified by distinctive DNA methylation patterns in order to further assess these cases. In addition to their distinctive epigenetic profile, the first analysis of the little molecular data did not reveal any recurring or distinguishing changes. Thus, further molecular studies were carried out.
Recently, IDH2 mutations or changes to the switch/sucrose non-fermentable (SWI/SNF) complex that cause SMARCB1 or SMARCA4 deficiency have been found to occur frequently in SNUCs. The present description of SNUC as a single entity is being called into question by these diverse molecular patterns, as well as the sporadic immunohistochemical and morphological similarities to neuroendocrine cancer.
The overall protein expression patterns for NEC-like IDH2 and NEC-like SMARCA4/ARID1A tumors were comparable in mass spectrometry-based proteomics. Observing the upregulation of multiple proteins that are unique to neurons or diffuse neuroendocrine system cells in both groups and gene set enrichment analysis also revealed a strong resemblance to neuroendocrine cells.
Clinical relevance
Disease-specific survival between the four SNUC classes is evaluated to further assess the clinical significance of the DNA methylation-based classes. When compared to instances from the NEC-like IDH2 (p = 0.012), NEC-like SMARCA4/ARID1A (p 0.001), or ACC class (p = 0.004), SMARCB1 class tumors had significantly lower disease-specific survival. The SMARCA4/ARID1A group, which resembles an NEC, had the highest chances of survival.
Epigenetic classes are defined using t-SNE and hierarchical clustering. The position of individual data points might fluctuate throughout successive iterations and greatly depends on chosen parameters and the makeup of the cohort, making t-SNE an unreliable method for classifying new instances. The information from the reference set is employed to create a machine learning algorithm that categorizes a given sample into one of the DNA methylation classes, allowing for easy and quick categorization of raw DNA methylation data in a diagnostic environment.
Support vector machine and random forest machine learning methods are examined, with the latter being the current gold standard for DNA methylation-based tumor classification, to determine the most effective method for this classification job.
Explanation
This paper provides a machine-learning technique for the reliable categorization of these diagnostically problematic tumors and a collection of DNA methylation patterns from a heterogeneous cohort of sinonasal cancers. Tumors with the SNUC shape are not as undifferentiated as their present name indicates but rather comprise four separate molecularly distinct entities combining DNA methylation profiling, DNA sequencing, copy number analysis, and mass spectrometry-based proteomics.
DNA methylation categorization was performed on a group of sinonasal tumors and related neoplasms that fall under the clinically relevant range. According to other research from related fields, the majority of recognized tumor types exhibit distinctive DNA methylation signatures that can be used to classify and differentiate them.
This might be explained by a number of factors
First, some of the omitted cases had diagnoses for which there were not enough instances to establish a distinct and stable class (e.g., biphenotypic sinonasal sarcoma). If more examples can be gathered, these items could be included in the next iteration of the classifier.
Second, technical differences between various tests or array qualities could contribute to somewhat varied DNA methylation patterns.
Thirdly, some of these instances may also have non-sinonasal origins, such as advanced malignancies from nearby anatomical areas (such as tumors that originated in the palate or brain) that continuously infiltrate sinonasal organs or distant metastases from an unidentified initial location. As they did not belong to a stable epigenetic class, these tumors were removed from further analysis, and none of them were used for the creation of the classifier.
Lastly, some instances of these non-clustering samples could relate to previously unknown, even rarer tumor classifications that need more research.
Investigating various DNA methylation signatures and molecular changes in the group of SNUCs that is diagnostically very difficult was another focus of this investigation. The findings suggest that the group of tumors now referred to as SNUCs likely consists of at least four discrete molecular classes, each of which has a unique genetic driver and a unique clinical outcome.
Constraints of the Study
- The gene body included the majority of the important CpGs. It is difficult to assess the regulatory impact of DNA methylation in these areas since it is not well known.
- More sensitive techniques like data-independent acquisition (DIA) or tandem mass tag (TMT) labeling, as well as phosphoproteomic profiling, might be beneficial for mechanistic investigations concentrating on less common signaling pathway components. Therefore, more research must be conducted.
- A central histological evaluation of the study’s case samples has not been conducted. Due to varying levels of experience in the diagnosis of sinonasal malignancies, the quality of the provided conventional diagnoses may vary amongst the institutions that offer them.
In conclusion, because there was very little information on additional outcome-associated clinical characteristics, including local tumor stage or metastatic stage, the outcome analysis should be taken with care. A DNA methylation-based method has been discussed in this study that may be a valuable diagnostic aid for sinonasal malignancies, avoiding misclassifications and assisting the workup of difficult patients. Additionally, by using SNUC morphology, the molecular heterogeneity of malignancies is better comprehended.
Article Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Riya Vishwakarma is a consulting content writing intern at CBIRT. Currently, she's pursuing a Master's in Biotechnology from Govt. VYT PG Autonomous College, Chhattisgarh. With a steep inclination towards research, she is techno-savvy with a sound interest in content writing and digital handling. She has dedicated three years as a writer and gained experience in literary writing as well as counting many such years ahead.