Are you aware of the perception of a genomic language model? If not, then it’s worth mentioning that researchers at Harvard, Cornell University, and the Massachusetts Institute of Technology developed gLM, a Genomic Language Model, which is trained on enormous amounts of metagenomic data to interpret gene contextual representations. Using pLM embeddings and transformer architecture, gLM learns gene functional semantics and regulatory syntax. It predicts gene functions and co-regulation patterns, with potential applications in numerous fields, including biotechnology.
Decoding the Genomic Language
At the heart of genomic language models is a thorough grasp of life’s crucial building blocks: DNA, RNA, and proteins. These models are proficient in massive volumes of genomic data, which allows them to understand the complicated patterns and correlations that control genetic sequences.
Genomic language models excel at untying complicated links among genes and their regulatory syntax, much as language models like GPT (Generative Pre-training Transformer) have reformed natural language processing by capturing the intricacies of human language. Through extensive training on unlabelled metagenomic sequences, gLM expertly navigates different regions of multi-gene sequences, revealing the subtle semantics and regulatory signals inherent in genomic data.
Orchestrating the Biological Symphony
Proteins, the cell’s workhorses, seldom function independently. Instead, they interact together in complex networks that are controlled by genetic constitution. Genomes regulate protein expression and affect their interactions. Understanding these patterns of co-regulation is critical for understanding the intricacies of biological systems.
While their model was mainly trained on bacterial, archaeal, and viral genomes, applying it to eukaryotic genomes, particularly those with large intergenic sections, demands more investigation.
One of the most tempting elements of transformer-based language models is their capacity to transfer learning and fine-tuning. So, researchers unveiled some of gLM’s abilities for learning higher-order biological information from genomic sequences, such as gene function and regulation. This emphasizes the necessity of contextualization in biological data modeling, particularly as we grow. Furthermore, they demonstrate gLM’s capacity to explicate context dependency in activities spanning structural and sequence homology, as demonstrated by AAA+ regulators.
Methodology Used
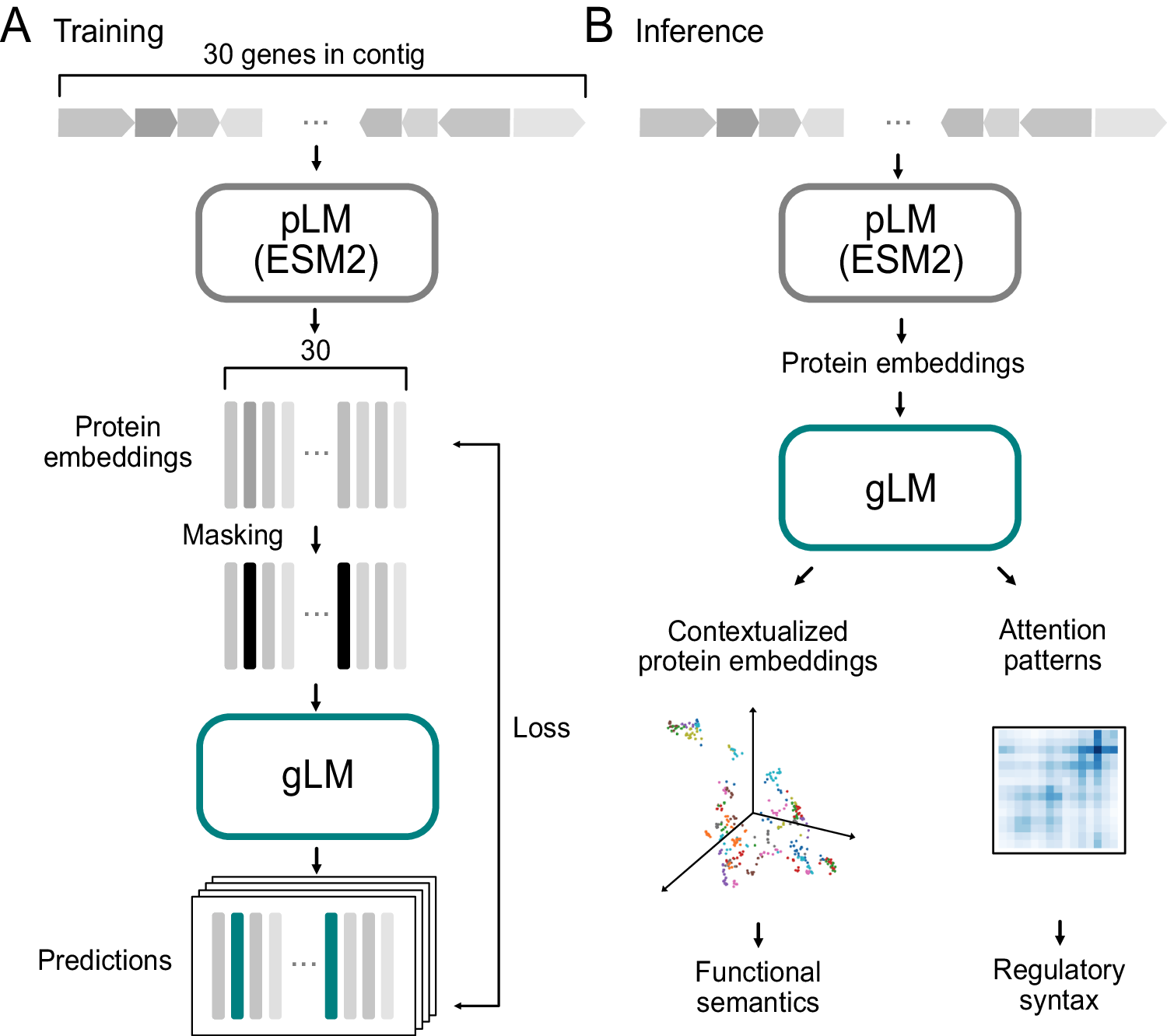
The technique used in the observation of genomic language fashions predicting protein co-regulation and characteristics involves numerous key steps:
Data Collection: The researchers accumulated tens of millions of unlabelled metagenomic sequences from databases, including MGnify, to train the genomic language version.
Model Architecture: They look at applying a 19-layer transformer version for the tutoring of the genomic language model. Transformers are neural community architectures regarded for their ability to seize lengthy-variety dependencies in sequential information.
Training Objective: The model becomes trained with the use of a masked language modeling objective, in which a fragment of the entered gene sequences is masked, and the version is tasked with reconstructing the original sequences. This technique helps the model scrutinize the semantics and syntax of genomic sequences.
Feature Illustration: Each gene in a genomic collection is denoted through a 1280 function vector, taking pictures of contextual statistics & relationships inside the gene series.
Contextualized Protein Embeddings: The observation fixated on producing contextualized protein embeddings, which encode relational residences and structural information of proteins. These embeddings are important for shooting the purposeful relationships among genes.
Attention Mechanisms: The genomic language model applied attention mechanisms to the recognition of extraordinary elements of the gene sequences, permitting the version to study dependencies and relations between genes.
Evaluation: The overall performance of the genomic language model is estimated on numerous downstream obligations, such as enzyme characteristic prediction, operon prediction, paralog matching, and contig taxonomy prediction, to evaluate its capability to predict gene features and co-regulation accurately.
An Overview of the Genomic Language Model (gLM)
The study provided fascinating insights into the genomic language model (gLM), notably its capacity to extract contextual information about enzyme performance. Notably, the first six layers of gLM were shown to encode a large amount of contextual knowledge, allowing for protein function prediction with up to 24.4% accuracy. Combining this contextual information with the original protein language model (pLM) data resulted in a substantial improvement, raising accuracy to 51.6% in the first hidden layer, a 4.6% gain over context-free pLM predictions.
Additionally, gLM embeddings were revealed to have unique information that was not captured by pLM embeddings, demonstrating that the model can capably detect extra functional and regulatory linkages. Remarkably, the study observed a decrease in the expressivity of enzyme function information with deeper gLM layers, which was consistent.
Importantly, the gLM representations showed high relationships with biological function and relational features similar to semantic information. Clustering evaluations confirmed the superiority of gLM embeddings over contig-averaged pLM embeddings, highlighting the model’s potential to capture subtle representations that go beyond basic baseline methods.
Looking Ahead
Numerous important areas for future development appear:
- Exploiting the transformer architecture’s efficient scaling capabilities has the potential to improve model performance as parameter and training dataset sizes rise.
- Future model iterations might include raw residue or codon-level embeddings to better capture residue-to-residue co-evolutionary interactions and synonymous mutation effects.
- Increasing the reconstruction of masked protein embeddings using generative techniques may improve prediction accuracy and generalizability for formerly unexplored datasets.
- Integrating non-protein modalities, such as non-coding regulatory components, may improve gLM’s representation of biological sequence data and allow for learning of protein function and regulation based on other modalities.
Concluding Remarks
The work on genomic language models for predicting protein co-regulation and function constitutes a big step in advancing computational biology. Researchers may now disclose hidden relations among genes and their genomic settings with unparalleled precision and complexity thanks to machine learning and deep neural networks. As we continue to study the huge vastness of biological data, genomic language models remain a beacon of hope for unveiling the secrets of life itself.
Article Source: Reference Paper | Training and inference code and analysis scripts are available on GitHub and Zenodo.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.