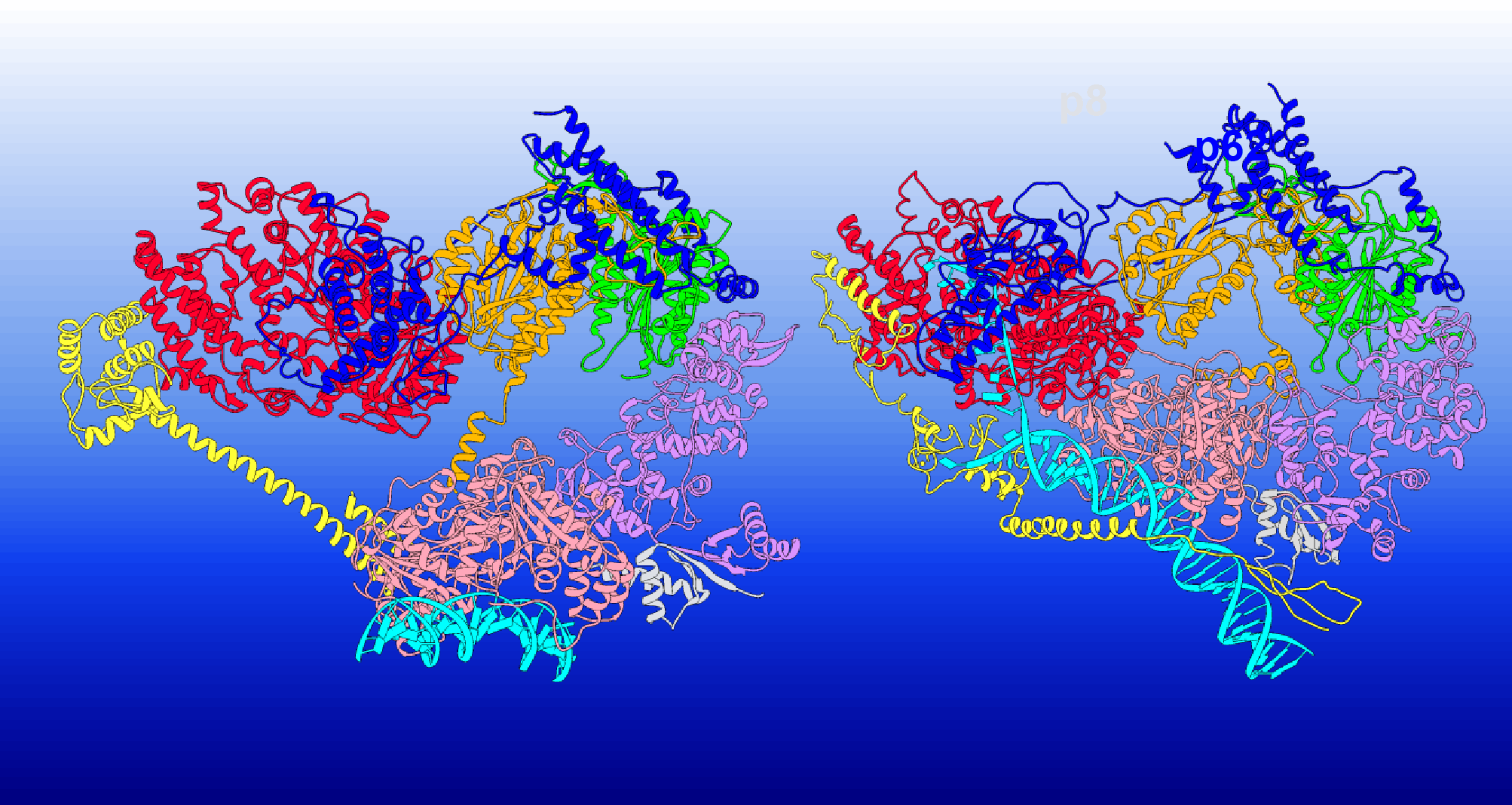
Scientists from Georgia State University, The University of Texas, and Lawrence Berkeley National Laboratory, USA, built models of transcription factor IIH (TFIIH) using cryo-electron microscopy (cryo-EM) to comprehend how it smoothly transitions between performing two vital yet vastly different functions, transcription initiation and nucleotide excision repair (NER). They employed simulations and graph-theoretical analysis and found that its two subunits, XPB and XPD, mediate conformational changes in TFIIH, enabling it to switch between transcription initiation and NER while ensuring that it performs only one function at a time. Another key observation they made was that diseases caused by TFIIH mutations can be grouped into distinct classes based on the interfaces of the constituents of TFIIH they affect.
The Fascinating Dynamism of TFIIH
TFIIH is a large and dynamic horse-shoe-shaped protein assembly consisting of ten subunits. The subunits include seven core ones (XPB, XPD, p62, p52, p44, p34, p8) and three CDK-activating kinase subunits (CDK7, Cyclin-H, MAT1). TFIIH is imperative to both transcription (enzymatic synthesis of RNAs from DNA templates), preinitiation complex (PIC), and nucleotide excision repair (removal of wide arrays of DNA lesions occurring due to UV radiation, reactive oxygen species, environmental and chemical mutagens), also called NER.
In transcription, TFIIH is a part of the assembly responsible for unwinding the DNA duplex and pushing it toward the RNA polymerase active site. TFIIH is basically the centerpiece of NER, as it assembles, coordinates, and regulates the NER machinery. And defects in NER are strongly associated with human genetic diseases such as Cockayne Syndrome, Xeroderma Pigmentosum, etc.
The most intriguing aspect is how TFIIH, a single protein assembly, is able to play a role in regulating two widely different genomic processes. Well, the answer is hidden in its modular architecture. Its architecture grants it such adaptability that it can bind to diverse protein partners. TFIIH can effortlessly transition from its apo state or unbound state (apo-TFIIH) to the holo state (holo-PIC) and further to the NER complex (NER-TFIIH).
However, the mechanisms underlying conformational switching and functional versatility are still unclear.
Uncovering the Differences Among TFIIH’s Three Conformations
Researcher Ivaylo Ivanov and his team conducted meticulous comparative analyses of the structure as well as dynamics of the three conformations of TFIIH, apo-TFIIH, holo-PIC, and NER-TFIIH, in order to get clarity on the mechanism behind its conformational transitioning and functional versatility. The team implemented cryo-EM to create three-dimensional images of the three different conformations of TFIIH.
The TFIIH/XPA/DNA cryo-EM structure was critical for delving deeper into the NER machinery (XPA is a protein involved in the repair of DNA damage). Even though the core subunits of the TFIIH could be modeled accurately into the cryo-EM density, some flexible structural elements, such as the C- and N-terminal ends of XPA and the whole p62 subunit, were not well-resolved. To complete the model, the missing elements were traced in the original EM density and built de novo.
The three models were then subjected to molecular dynamics (MD) simulations to study the physical movements of atoms and molecules in the model. The translocation of DNA by the two ATPase subunits of TFIIH, XPB, and XPD was also modeled with the help of chain-of-replicas path optimization methods. A method called difference contact network analysis (dCNA) was also applied in the three models to compare the interactions between proteins in the three complexes.
From all these methods administered in the study, some key differences among the three complexes were observed, such as:
- Apo-TFIIH and PIC encompass all ten subunits of TFIIH, while the NER complex only includes the seven core subunits.
- In Apo-TFIIH and PIC, XPB and XPD are spaced out, while in the NER complex, they are adjacent.
- In Apo-TFIIH and PIC, MAT1 (a protein involved in the transcription of DNA) is positioned between XPB and XPD, while in the NER complex, it is displaced by XPA and XPD ssDNA binding.
- In Apo-TFIIH and PIC, DNA is directed away from XPD, while in the NER complex, it is directed towards XPD.
- In Apo-TFIIH and PIC, XPA does not play a significant role, while in the NER complex, it helps to clamp DNA onto XPB and to separate the DNA strands.
XPB and XPD: Two proteins, two distinct DNA translocation mechanisms
The unwinding of DNA in NER is reliant on two subunits of TFIIH, XPB, and XPD. Yet, how they cooperate in unwinding DNA and scanning lesions during NER remains unclear. The researchers developed individual models of the two subunits to understand their mechanisms and study their behavior. They used chain-of-replicas path optimization, especially the partial nudged elastic band method (PNEB), to calculate the minimum free energy paths (MFEP) for the movement of DNA through XPB and XPD. These paths represented the entire process of DNA movement and also disclosed the main motions of XPB and XPD that drive the forward movement of DNA as the nucleotide state changes.
XPB interacts with double-stranded DNA using its RecA1 and RecA2 domains. These domains rotate in opposite directions, causing the DNA to rotate and move forward along the XPB groove. The movement occurs in small steps, and the ATPase domains of XPB open and close during the process. NTE and DRD domains form a latch that supports the ATPase core from the opposite side of the DNA-binding groove, reinforcing the connection between the ATPase domains, which would otherwise be held together only by a single flexible linker. In the TFIIH complex, this latch is part of a collar structure that helps direct the motions of XPB and promotes the forward movement of DNA.
XPD moves along single-stranded DNA through a narrow channel formed by its structural domains, RecA1, RecA2, iron-sulfur domain (Fe-S), and Arch domain. The channel has two constricted regions. The first constriction is located between the Arch and Fe-S domains and involves the opening and closing motions of the Arch and Fe-S domains. This motion is functionally imperative because if ssDNA gets blocked between the Arch and Fe-S domains, it could indicate the presence of a bulky lesion. The second constriction is on the opposite side of the XPD DNA-binding cleft. Unlike the first constriction, it does not open up and ensures a controlled and regulated movement of nucleotides through XPD’s channel, preventing multiple nucleotides from passing through simultaneously.
Video Source: Oak Ridge National Laboratory
The Enigma of TFIIH Finally Figured Out!
From this study, the many intricacies of TFIIH were discovered. The researchers found that the conformation of TFIIH changes depending on the functions it performs. The three different forms of TFIIH demonstrated different flexibilities, and their flexibility depends on the spacing between XPB and XPD. In holo-PIC, the spacing is the largest, and hence, it is the most flexible out of the three forms. Apo-TFIIH shows intermediate flexibility, and NER-TFIIH has the least flexibility.
Using a network analysis method, the researchers mapped the protein’s structural relationships and looked at how different protein parts interact.
They inferred that during the transition from one form to another, the interface between XPB and XPD changes. During the process of transcription initiation, when TFIIH is in the holo-PIC form, XPB acts as the active component unwinding DNA while XPD is directed away from DNA, and its DNA binding groove is blocked by another subunit of TFIIH called p62. In contrast, during NER, XPD assumes the role of the active component, and XPB serves a purely structural role.
This study also sheds light on the aspect of mutations in TFIIH. Mutations in TFIIH can negatively impact NER and cause genetic diseases, and the researchers found that most disease-causing mutations are located at the interfaces between different parts of TFIIH, especially those between XPB and XPD subunits. For example, mutations associated with xeroderma pigmentosum and Cockayne syndrome are located in XPD, and mutations causing Trichothiodystrophy are found in XPB.
Conclusion
TFIIH is responsible for controlling two highly crucial life processes, and any defect in it leads to disastrous consequences in the form of dangerous diseases. Therefore, understanding the workings of TFIIH is the key to potentially reversing or countering the effects of mutations. Now that the mystery of TFIIH’s functional versatility is beginning to unravel, it is highly likely that we can delve even deeper into its workings and find reliable therapeutic practices to cure deadly genetic diseases in the future.
Article Source: Reference Paper
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.