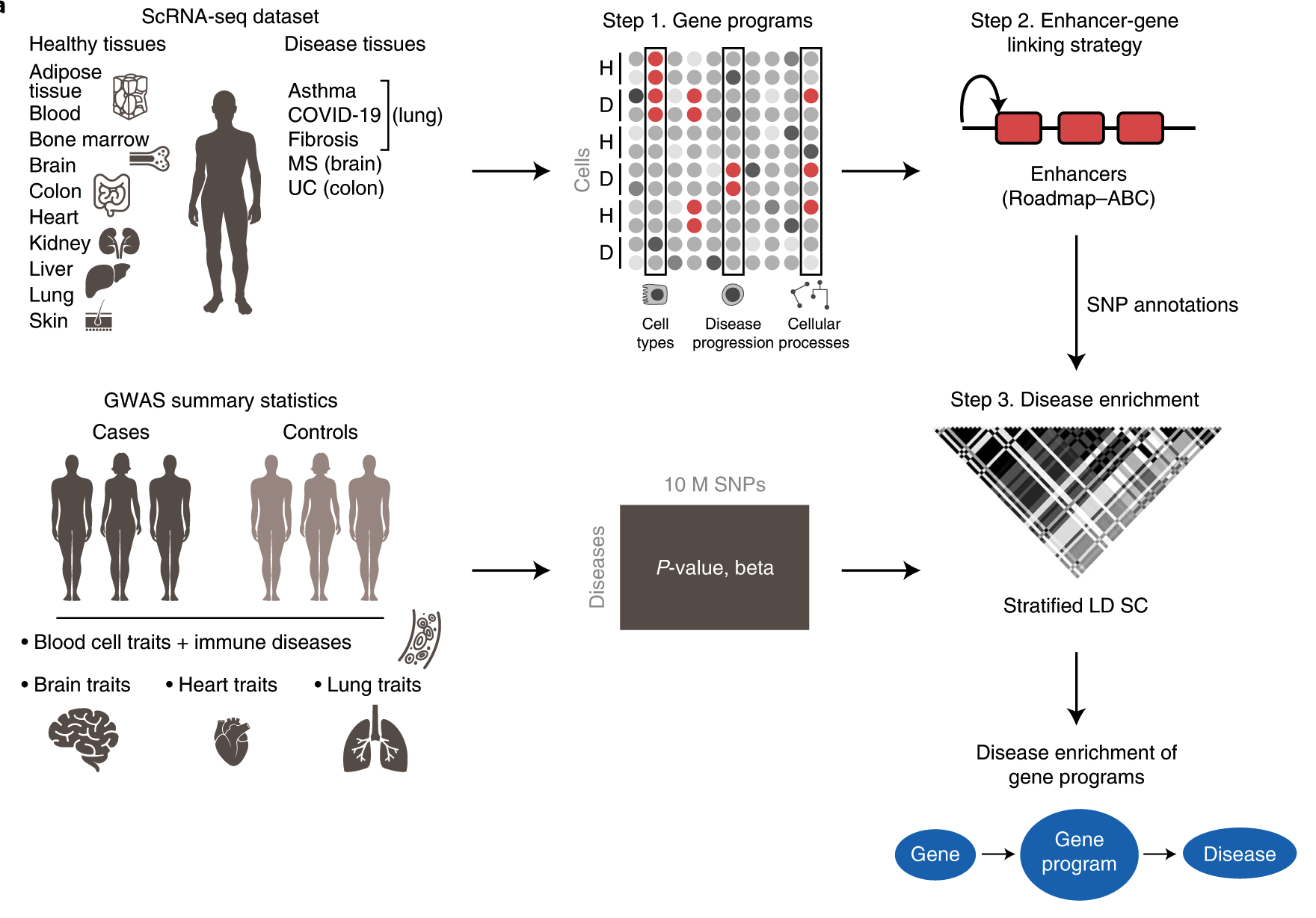
A brand-new framework called sc-linker has been proposed by researchers from the Broad Institute of MIT and Harvard that connects human illness and phenotypes to different cell types and physiological processes. This is done by integrating Genome-Wide Association Studies summary statistics, epigenomic data, and single-cell RNA (scRNAseq) sequence data from multiple tissues, cells, diseases, and individuals.
Genome-wide association studies (GWAS) entail quickly scanning markers throughout the whole genomes or DNA sets of several individuals to identify genetic variants linked to a specific disease manifestation. When a novel genetic link is discovered, researchers can utilize that knowledge to improve illness detection, treatment, and prevention methods. While GWAS has effectively found several variants associated with illness, little is known about the biological processes driven by these variants. This is because nearly 95% of the disease risk variant lie in the non-coding regions of the genome.
Single-cell RNA seq provides transcriptional profiling of thousands of individual cells. This approach provides a more detailed understanding of cellular distinctions and the relationship between the cell and its surroundings. Data collected from various single-cell omics methods can allow the construction of multiple gene programs and relate it with particular cells or tissue and the plausible disease signatures.
It might be deceptive to concentrate just on a single kind of cell or tissue in bringing about any disease phenotype; thus, it is necessary to connect genomic and single-cell RNAseq data to many cells, tissues, and even organs. The current study by Jagadeesh et al. reports a novel integrated framework named sc-linker to relate human diseases to cell processes using GWAS results and sc-RNA seq data.
Findings
The authors gathered human scRNAseq data from 17 separate datasets to create gene programs. Gene programs are characterized by unique cell types, processes, and disease-dependent signatures. The genes were then linked to Single Nucleotide Polymorphisms, and using GWAS data from a wide range of complex illnesses and characteristics, the genes that were shown to be significantly involved were reported and further examined. This work uses the S-LDSC software to process GWAS data as well as S-LDSC software and MAGMA v.1.08 for post hoc analysis.
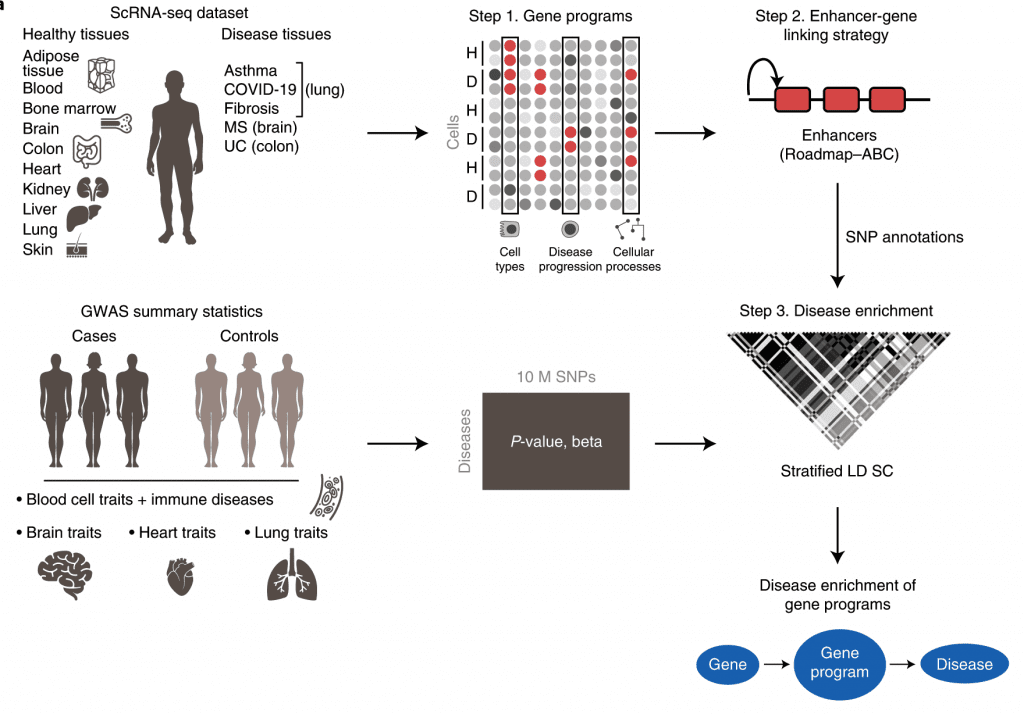
Image Source: https://doi.org/10.1038/s41588-022-01187-9
Researchers then went on to investigate the robustness of sc-linker to various diseases. Nine cell-type programs and twelve cell-process programs from the RNA data obtained from a healthy brain versus RNA data obtained from 11 neurological illnesses were analyzed. It was observed that there were enriched GABA-ergic neurons underpinning Major Depressive Disorder (MDD) and Body Mass Index (BMI). Glutamatergic neuron enrichment was observed in insomnia, schizophrenia, and intelligence. With current therapies for MDD targeting the neurotransmitter serotonin, the GABA-ergic neuron enrichment showcases that there are other plausible, targettable pathways in MDD, still unexplored.
The authors also examined Ulcerative Colitis (UC) and Irritable bowel syndrome (IBS) GWAS data with healthy cell type. The gene programs responsible for ulcer disease signatures and ulcer-driven cellular pathways were collected from the scRNAseq data of both
healthy and inflamed colon cells. This analysis showed an enrichment of MHC-II antigen presentation, complement cascade, and EGFR signaling pathways in healthy and diseased intestinal cells. The sc-linker highlighted an increased number of M-cells in ulcers, genetically, mutations in FERMT1, which is known to cause Kindler’s syndrome that resembles Ulcerative Colitis and IBD-like symptoms. The FERMT1 mutations are a driving force in bringing about M-cell-dependent disease.
GWAS data for asthma, covid-19 were linked with scRNAseq data from healthy tissues and tissues with asthma or covid-19. Sc-linker highlighted the significant enrichment of T-cells and MAPK pathways driving covid-19, idiopathic pulmonary fibrosis (IPF), and asthma.
Major Limitation Of The Study: The cell types, process & disease states covered in the work are not exhaustive enough; as the power of sequencing grows, the cell types and granular cell states also grow.
Final Thoughts
The article demonstrates new unexplored biological information gained by linking cell types with genomic disease signals. Novel notable enrichments identified by sc-linker ensure that the framework has the potential to identify new knowledge in disease mechanisms, although this needs to be confirmed experimentally.
In the future, connecting gene programs and disease mechanisms can help in designing further tests. With increasing success in phenome-wide association studies and integration with epigenomic data, sc-linker will continue to help identify broad-range disease phenotypes.
Article Source: Reference Paper | Web Interface
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Shwetha is a consulting scientific content writing intern at CBIRT. She has completed her Master’s in biotechnology at the Indian Institute of Technology, Hyderabad, with nearly two years of research experience in cellular biology and cell signaling. She is passionate about science communication, cancer biology, and everything that strikes her curiosity!