Scientists from Yale University developed a novel private blockchain network by employing layered database indexing for the storage of reference-aligned next-generation sequencing reads and individual genetic variants.
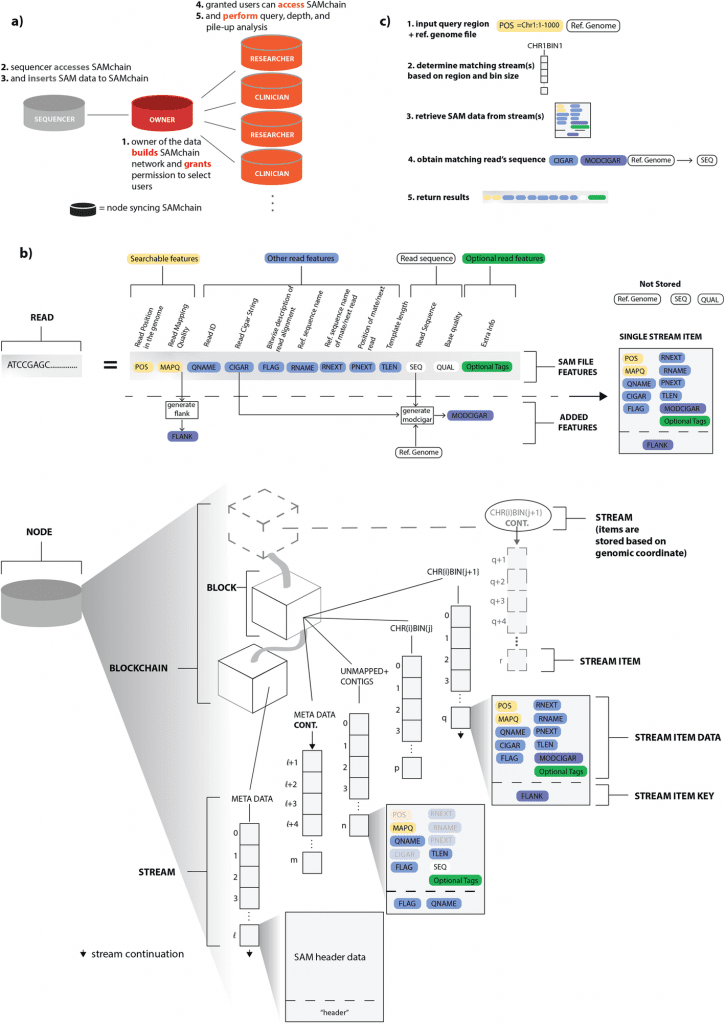
Image Source: https://doi.org/10.1186/s13059-022-02699-7
Major initiatives are being made to integrate genome sequencing into clinical practice regularly.
Finding workable solutions for data ownership and integrity is a significant hurdle. Blockchain offers answers to these problems in other fields, like finance.
However, the inability to store massive amounts of data on-chain, the sluggish transaction speeds, and the querying restrictions prevent its widespread application in genomics.
The scientists created a private blockchain network to store genomic variants and reference-aligned reads in order to get around these obstacles.
To access the data quickly and do analysis, it employs nested database indexing and a supporting toolset.
Genome Sequencing in Routine Clinical Care
The time is coming closer when genomic data will drive healthcare and biomedical research as a result of recent developments in personalized medicine, which have led to an increase in the number of people who are willing to sequence their genomes for disease-risk assessments and ancestry analysis.
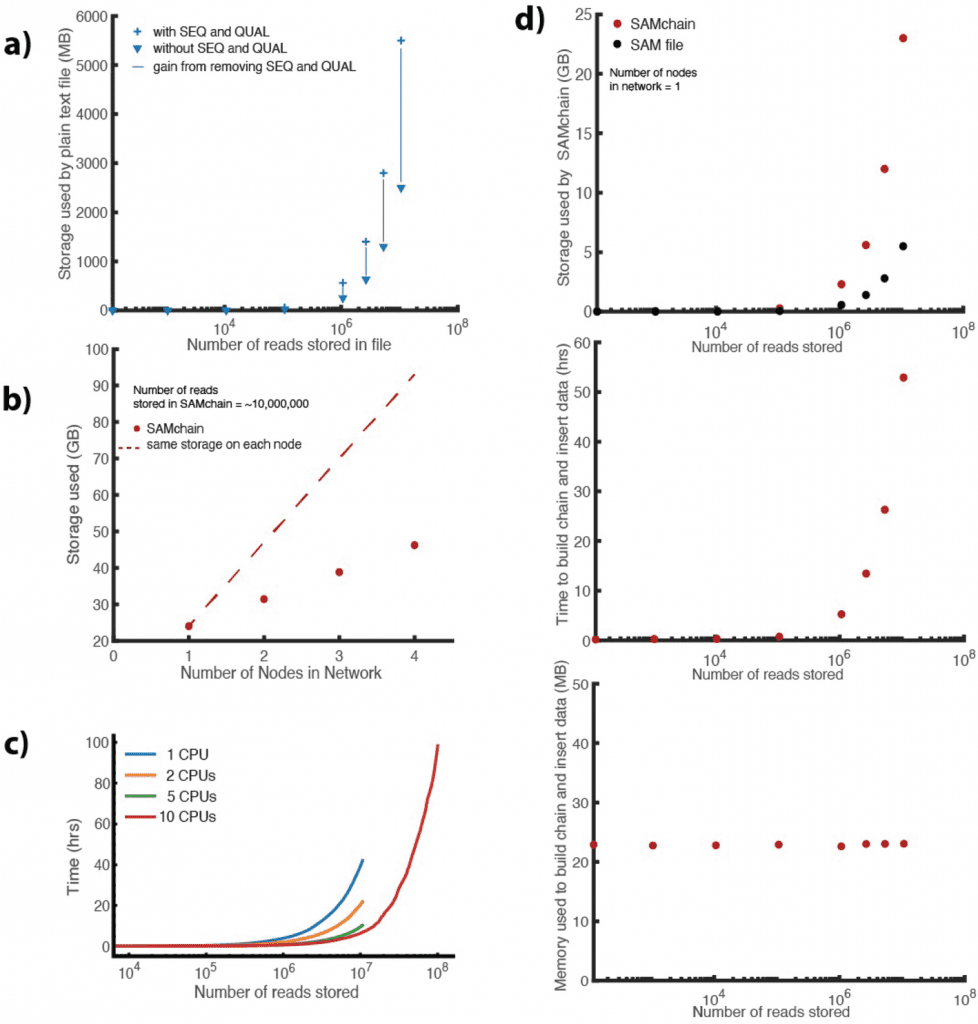
Image Source: https://doi.org/10.1186/s13059-022-02699-7
Genome sequencing will eventually become a standard component of clinical care, and the number of human genomes that have been sequenced will only increase, given the enormous interest in understanding one’s genomic data and the potential of genomic data for furthering biomedical research.
The rise in Human-Derived Functional Genomic Datasets
Data ownership issues have come up with the increase in genetic testing in hospitals and through direct-to-consumer businesses.
Data ownership, which denotes both power and control, is the possession and responsibility of the information contained in the data.
The ability to access, create, modify, package, reap benefits from, sell, or remove information is referred to as control of the information. It also includes the right to grant others these access privileges.
Individuals should ideally have control of and access to their data. This gives people control over their own genetic data’s right to sharing (privacy) and method of sharing (security).
However, the infrastructure needed for storage and analysis is missing. Because of this, there is no technical way to ensure the provenance and security of this extremely private, personal data, allowing data owners to advance science and medicine while still maintaining the security of their data.
Furthermore, difficulties with data ownership are not just applicable to genomic data at the personal level.
Human-derived functional genomic databases are expanding, thus, it is important to pay attention to who owns and manages these datasets.
Blockchain for Storage and Analysis of Genome
The potential of blockchain technologies is gaining more and more attention. Discussions have recently focussed on how blockchain technology could alter the world’s currency by simultaneously addressing several problems with the current financial systems (e.g., transaction speed, security, and removal of middle man).
Using non-fungible tokens (NFTs) as an illustration, valuable assets are increasingly being deployed on open or hybrid blockchains.
Additionally, a variety of industries, including supply chain management and the media and entertainment industry, are using blockchain technology.
The capabilities of this technology in genomics are obviously of interest, as blockchain technology may establish verified and public proof of data ownership.
But no one has yet worked out how to use a blockchain to store a significant amount of data, like the read stack seen in genome sequencing.
This is because blockchain presents technical challenges for analyzing and storing massive amounts of data. In the given study, the scientists employ a method to store and examine a genome in a blockchain for the first time.
Blockchain-based solutions for the exchange of genomic data are provided by numerous burgeoning biotech firms.
Many blockchain genomics organizations store the genomic data elsewhere, such as in Blockstack or the InterPlanetary File System (IPFS), and use blockchains as a log-keeping infrastructure due to the technical difficulties associated with storing genomes in blockchain architecture.
In order to provide a robust system for file storage and sharing, IPFS is a decentralized file system that uses a distributed hash table for quick access to the shared files.
Converting a Private Blockchain into a Consortium Blockchain
Due to storage redundancy and network verification mechanisms, blockchain applications demand more computing power and storage space than centralized database applications.
When storing and retrieving data, the decentralized system also causes a larger latency (delay in data transfer).
Additionally, the blockchain network’s transactions take time to publish data to the chain since they need to undergo cryptographic consensus verification.
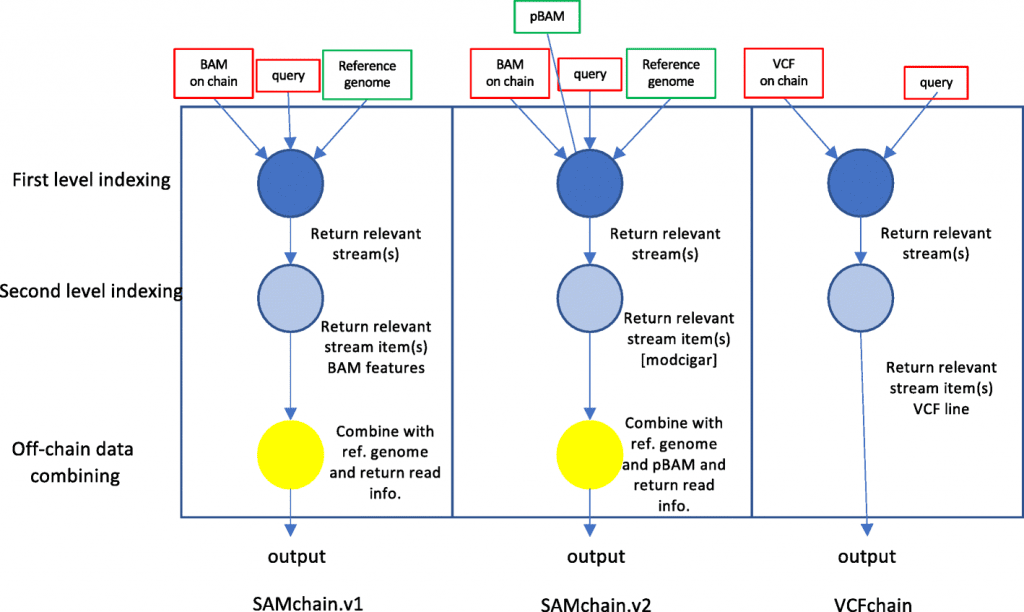
Image Source: https://doi.org/10.1186/s13059-022-02699-7
The researchers suggest that the issues associated with data ownership and control as well as technical challenges can be resolved by a well-designed private blockchain network.
Additionally, it can maintain the data’s security and integrity. When developing solutions for storage and analysis, genomic data’s integrity and security must be given top consideration because it is becoming more and more crucial to our comprehension of human health and disease.
Future health care and research integrity issues could be caused by the corruption, modification, or loss of personal genomes.
Personal genomic data storage would be implemented in a way that:
- Protected against loss and manipulation;
- Allowed doctors and biomedical researchers proper access; and
- Gave individuals authority over their genomic data.
Despite the lack of mathematical assurances at the level of validation, private blockchains use cryptographic protections and blockchain-specific data structures (such as Merkle trees) to ensure that invalid transactions are not added to the chain.
Although proof-of-work is not employed, alternative consensus processes are used to verify transactions.
To demonstrate their benefits, private blockchain networks can be compared to distributed databases. Blockchain architecture has a number of benefits.
In a nutshell, the benefits of blockchain include decentralized database management, immutability (as a result of a new block’s hash including the details of blocks that came before it), data provenance, robustness, security, and privacy.
Further enhancing its security, a private blockchain may be changed into a consortium blockchain where consensus is managed by a pre-selected group of nodes (i.e., proof of work in a limited way).
Presentation from the Study
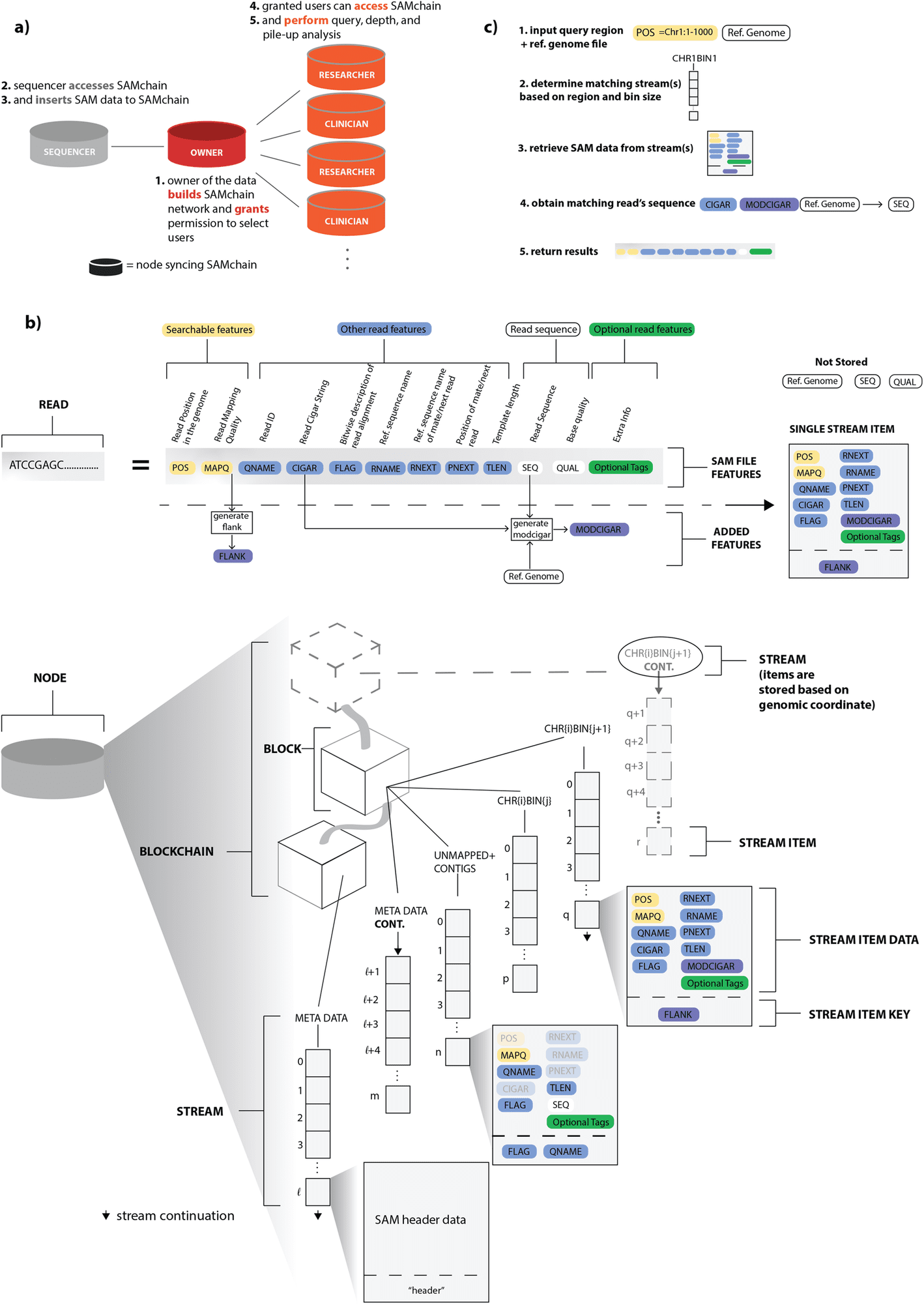
In the given work, scientists propose the first private blockchain network that is open-source and proof-of-concept, allowing for efficient storage and retrieval of raw genomic reads and individual genetic variants.
With the open-source blockchain API MultiChain, they created unique data structures based on layered database indexing, file-format alterations, and compression methods to address the abovementioned issues.
They utilized their “data stream” function, which enables users to build several key-value, time-series, or identity databases for use in data sharing, time-stamping, and encrypted archiving.
Two modules are available:
- The first module is a small piece of software that enables users to quickly save and search VCF files (VCFChain and VCFtool)
- The second module, which uses more resources, enables users to establish a chain and insert SAM data into it (SAMchain) as well as carry out a variety of tasks (SCtools), including querying, depth analysis, pile-ups for variant calling, and creating new SAM files and their derivatives (such as BAM and CRAM files).
The researchers concentrated on improving the data storage towards speedier querying/analysis using several levels of indexing schemes since the act of pushing the data into a blockchain is done just once while querying and analyzing the data is a continual process.
This contrasts with the present blockchain genomic data storage alternatives made available in the private sector, where the data storage is optimized, and no option is provided for data querying and analysis.
The Endpoint
The researchers envision a world where people build private blockchains to hold their genomes, which they then share with their healthcare providers and biological researchers.
Healthcare professionals and geneticists can stream or query patient genomes simply by virtue of intrinsic access control and security capabilities.
When a piece of data is encrypted on a shared server or in the cloud, for instance, there is a chance that it could become corrupted, especially if the data are very vast, like genetic data.
Additionally, conventional access-control methods like dbGAP or EGA exclude the study subject or patient from the decision-making process.
Their private blockchain network lowers the danger of data corruption, prevents unauthorized access to private data, and returns control to the data’s rightful owner.
Since immutability is a feature of blockchain, data cannot be changed accidentally or on purpose.
This is a crucial component of the system for both large-scale open genetic data and personal, sensitive data.
Large-scale open data, like that from the 1,000 Genomes Project or the Personal Genomes Project, are susceptible to corruption and tampering, especially in centralized data storage systems.
Furthermore, to further manage and audit access more effectively, the blockchain-based system can be integrated with blockchain-based data access auditing tools.
To the best of the scientists’ knowledge, their framework is the first open-source program that enables blockchain-based genomic data streaming and querying.
In comparison to the present biomedical blockchain applications, this is a significant improvement. Their approach may be expanded to store encrypted data in the data streams to solve privacy concerns. For instance, a lot of bioinformatics issues have lately been addressed using homomorphic encryption.
It allows for direct calculations of encrypted data on any shared server, even the public cloud, while guaranteeing mathematical privacy.
The data could even be homomorphically encrypted, enabling direct calculations on the encrypted data. However, this would result in increased storage and processing costs.
Blockchain is a revolutionary new technology that has addressed issues with ownership and integrity in various fields (e.g., finance and art).
However, the difficulty of storing massive amounts of data on a blockchain prevents its utility in genomics. In the given study, the scientists offer a way around these impediments.
The researchers created a novel private blockchain technology network employing layered database indexing to store reference-aligned next-generation sequencing reads and individual genetic variants.
The researchers also created tools for quickly obtaining and examining the on-chain data. By reducing the amount of data injected into the chain by utilizing reference-based data compression and indexing algorithms, they were able to overcome the difficulties associated with on-chain data storage.
Additionally, their tools offer open-source blockchain-based access and storage for sophisticated genomic analysis like a variant calling.
Article Source: Gürsoy, G., Brannon, C.M., Ni, E. et al. Storing and analyzing a genome on a blockchain. Genome Biol 23, 134 (2022). https://doi.org/10.1186/s13059-022-02699-7
Learn More About Bioinformatics:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Tanveen Kaur is a consulting intern at CBIRT, currently, she's pursuing post-graduation in Biotechnology from Shoolini University, Himachal Pradesh. Her interests primarily lay in researching the new advancements in the world of biotechnology and bioinformatics, having a dream of being one of the best researchers.