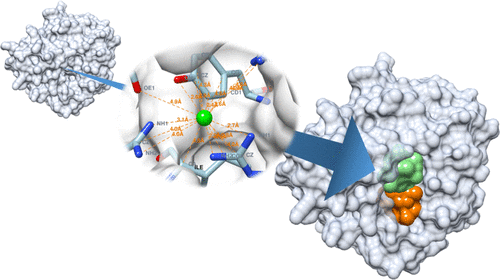
The identification of ligand-binding sites on the surface of a protein is a vital aspect of structure-based drug design. SiteRadar is a new algorithm for identifying ligand-binding sites on protein surfaces, which is important for structure-based drug design. It outperforms existing techniques, including machine learning algorithms, and can detect up to 74% of true ligand-binding sites. SiteRadar has the potential to be used in automated drug design algorithms, making it a promising solution for improving the efficiency and accuracy of the drug discovery process.
Limitations of existing methods and SiteRadar
Modern processes for drug design include ligand-based drug design (LBDD) and structure-based drug design (SBDD). SBDD is especially beneficial when the three-dimensional structure of a target protein is known, but information about possible ligands is unknown. SBDD begins with the examination of the target’s structure and the identification of a druggable pocket, which increases the potential for drug design. The costly and time-consuming biophysical approaches for discovering protein-ligand binding sites have prompted the development of computational methods. Existing computational techniques have limited predictive power, particularly for detecting shallow, allosteric, and covalent binding sites, as well as protein-protein interface areas.
SiteRadar is a computer program that detects protein-ligand-binding sites using a graph neural network. It depicts a protein structure as a graph and considers the distance between atoms and amino acid characteristics. SiteRadar was trained on a library of protein structures and can detect binding sites on the surface of a protein with precision. It outperforms previous approaches and can distinguish between various types of binding sites, making it suitable for automated drug development.
Comparison of SiteRadar with other models and different metrics
The study compared the performance of SiteRadar to that of Fpocket and PUResNet in finding protein binding sites. SiteRadar beat the other two approaches in terms of accurately recognizing real binding sites and producing fewer false positives. Yet, Fpocket and PUResNet matched the pocket form more precisely. In addition to having a better PC score than the other two approaches, SiteRadar has a lower DVO metric accuracy. Overall, SiteRadar demonstrated a greater capacity for discriminating when finding protein binding sites.
In addition to comparing SiteRadar to Fpocket and PUResNet, the study examined various combinations of the Agglomerative Clustering threshold and MeanShift bandwidth parameters. To evaluate SiteRadar’s performance under varying parameter values, the same metrics were computed for 17 other configurations using varied parameter values. This investigation attempted to optimize the SiteRadar parameter values and give users with recommendations for optimum outcomes.
Case studies
The SiteRadar algorithm was applied to 3D structures of pharmacological targets that were dissimilar to the training set in terms of amino acid sequence similarity. This was done to demonstrate the algorithm’s applicability to targets with no analogs in the training data set. The maximum sequence similarity to the training set did not exceed 43%, indicating that the targets were sufficiently different from the training set. Despite this dissimilarity, the SiteRadar algorithm was able to identify potential binding sites in these targets, demonstrating its broad applicability domain.
The surface of NADH oxidase
In this case study, the SiteRadar method was utilized to discover a binding site on the surface of NADH oxidase, which is notoriously challenging to detect using geometric and chemical information-driven techniques. SiteRadar (AAspecific) detected the binding site using chemical information and did not find any false-positive pockets. In contrast, SiteRadar (geometric) discovered just a partial binding site and an extra cavity around the binding site. This example illustrates the benefit of utilizing chemical information to identify solvent-exposed binding sites, especially when geometric techniques are insufficient.
Polycomb protein EED
Polycomb protein EED (Embryonic Ectoderm Development) is a member of the Polycomb group (PcG) of proteins that play an essential role in regulating gene expression during development. SiteRadar used two approaches here – AA-specific and geometric – to detect potential binding sites. Both models successfully identified an allosteric site and additional pockets, but the AA-specific model was more selective and focused on specific amino acids, while the geometric model covered adjacent areas more broadly.
Covalent ligand binding in the CREB-binding protein
Although not designed expressly for this purpose, both the geometric and AA-specific techniques effectively identified the covalent binding site and covered nearly all heavy atoms of the cocrystallized ligand, with the exception of the covalent warhead, which was exposed to the solvent. The results indicate that SiteRadar may be utilized to identify potential binding sites for small-molecule covalent inhibitors. In this instance, the AA-specific model offered a more precise match to the ligand’s form, albeit the difference between the two techniques was negligible.
PPI-binding site
Using both chemistry-driven and geometric models, the SiteRadar software was utilized to map replication protein A. The AA-specific model uncovered two distinct clusters, but the geometric model uncovered a single widespread binding site. SiteRadar was proven to be successful at identifying various ligand-binding sites, including PPI sites, and the choice of model is dependent on the shape and amino acid composition of the pocket.
Conclusion
SiteRadar is a precise technique for detecting protein-ligand binding sites that do not need data preprocessing or partial charge computation. It is applicable to a wide variety of binding sites, including non-conventional ones, and is especially valuable for computer-assisted drug design. Due to an appropriately balanced quantity of created pockets and a precise scoring mechanism, its accuracy is greater than alternative approaches such as Fpocket and PUResNet, which are not ideal for screening a large number of proteins. SiteRadar prefers to map pockets in a more selective manner, avoiding unnecessarily big binding sites and separating large anticipated pockets into many binding sites of suitable size.
Article Source: Reference Paper
Learn More:
Sejal is a consulting scientific writing intern at CBIRT. She is an undergraduate student of the Department of Biotechnology at the Indian Institute of Technology, Kharagpur. She is an avid reader, and her logical and analytical skills are an asset to any research organization.