Drug resistance can be countered only by interrupting the adaptive measures taken by the cells to deflect the drug action. Scientists from the School of Pharmacy and Pharmaceutical Sciences, University of Buffalo, performed an extensive study through proteomic analysis that identified key regulators driving the resistance against Gemcitabine, the principal chemotherapeutic against Pancreatic Adenocarcinoma. The study also mentions viable pathways and protein targets onto which therapeutic efforts could be directed to better improve the drug efficacy.
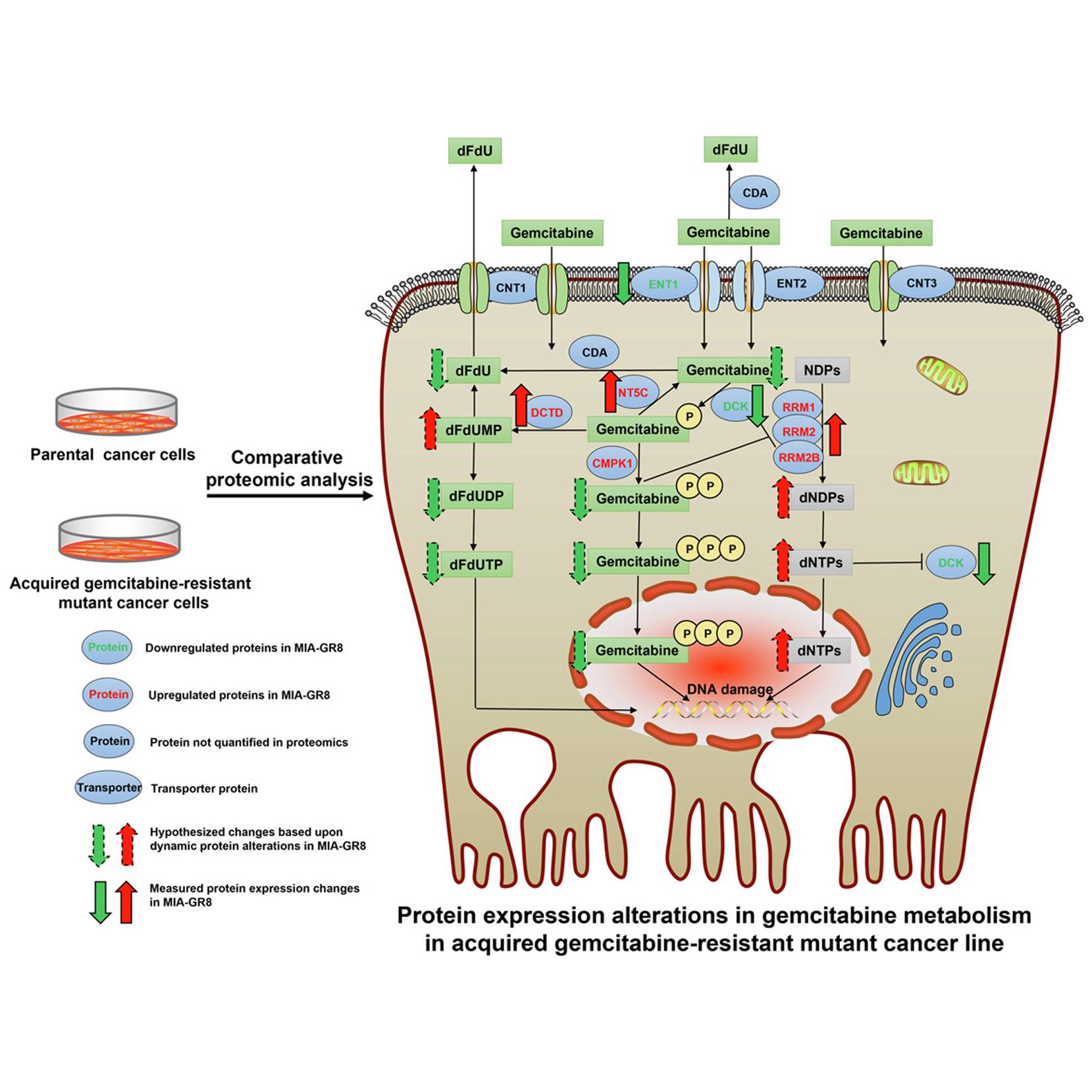
Gemcitabine, first marketed under the brand name Gemzar by Eli Lilly, is an antimetabolite capable of retarding tumor progression and promoting apoptosis by inhibiting DNA synthesis. On its uptake into the tumor cell, the compound is phosphorylated to form gemcitabine monophosphate, which acts as the precursor for its diphosphate and triphosphate forms. The diphosphate interacts and inhibits the ribonucleotide reductase (RNR) complex that maintains the nucleotide levels within the cell, while the triphosphate is a structural analog of deoxycytidine triphosphate(dCTP) and masquerades around as dCTP on its synthesis. In a nucleotide deficient state caused by gemcitabine diphosphate, it competes with dCTP for incorporation into the elongating DNA strand. On achieving this, it causes chain termination, ultimately dismantling the DNA synthesis machinery of the cell.
Considered one of the deadliest cancers with a five-year survival rate of 8%, pancreatic adenocarcinoma (PDAC) is still in dire need of effective treatments that can improve patient outcomes. Gemcitabine is appraised as a gold standard in exerting local control and improving survival rates in PDAC. But its effects are stunted by chemoresistance that arises within weeks of starting the treatment. To design therapeutic approaches that circumvent it, a thorough knowledge of key cellular regulators in drug response and resistance is imperative. On this note, the scientists at the University of Buffalo performed an extensive proteomic study by global quantification of proteins from PDAC cell lines using mass spectrometry to identify protein expression alterations favoring drug resistance.
GEMCITABINE – SENSITIVE AND – RESISTANT CELL LINES
MIAPaca-2, gemcitabine-sensitive PDAC cell lines, were exposed to continuously escalating levels of the drug until they developed the desired level of resistance against it. Of the nine gemcitabine-resistant clones developed from the parental cell lines, MIA-GR8 displayed the highest resistance and hence was chosen as the main protagonist for the study.
In the initial few analysis, MIA-GR8 cells exhibited slower growth both in in vitro and in vivo studies when compared to their parental cell lines. It is inferred as an adaptive mechanism to evade the drug action that is heavily based on targeting the cell cycle progression. In fact, in vitro studies identified around 60% of the cells stagnant at the G0/G1 phase of the cell cycle. In in vivo studies performed by implanting the cells into SCID mice, the MIA-GR8 tumor displayed slower progression that was unaffected by gemcitabine treatment. In addition, MIA-GR8 cells had increased expression of Zeb1 and Slug, markers of mesenchymal cells, revealing endothelial to mesenchymal transition (EMT) that imparts enhanced invasiveness and migration abilities.
PROTEOMICS ANALYSIS OF GEMCITABINE RESISTANCE
Proteomic analysis was performed through the IonStar pipeline, which was designed to handle MS1 ion-current data from mass spectrometry analysis in a way that allows for handling greater coverage depth (quantifiable proteins) across large sample counts with high quantitative accuracy. The use of this method allowed the authors to quantify around 6000 proteins across all study samples to identify the differentially expressed ones and thereby unearthing mechanisms conferring gemcitabine resistance. Protein quantifications were performed on Day 0 and Day 4 of culturing to monitor the protein expression patterns.
Annotation of differentially expressed proteins between gemcitabine-sensitive and resistant cells exposed the significant biological processes that aid in attaining drug resistance. Of which, the highest number of protein alterations assisted in lowering the energy production within the cell. A prominent contribution of it was through drug metabolic processes, which boast the highest upregulated protein of the study, RRM, with a log fold change of around 20.
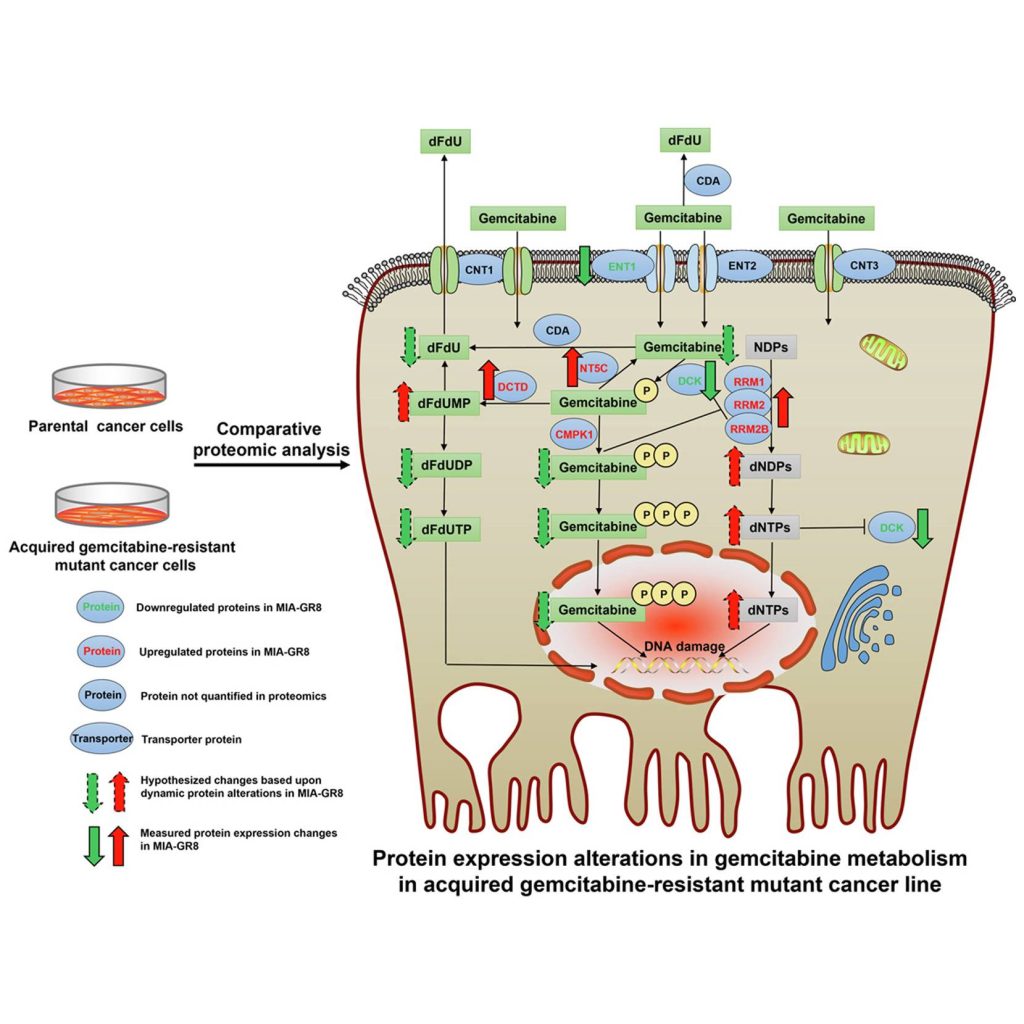
Image Source: https://doi.org/10.1016/j.mcpro.2022.100409
RRM1, along with RRM2 and RRM2B subunits, both of which showed upregulation in MIA-GR8, form the RNR complex that, as mentioned in the first paragraph assists in the nucleotide production and DNA repair leading to a subsequent reduction in gemcitabine incorporation into the DNA strands. RRM1’s role was further investigated by silencing the said gene in the MIAPaca-2 cell lines, which showed increased levels of apoptotic caspases, reduced cellular viability, decreased migration, and lowered expression of EMT markers. This observation was validated through clinical data where increased RRM1 expression inversely affected patient survival. Other upregulated drug metabolic proteins included those involved in the dephosphorylation of Gemcitabine into its inactive states that are subsequently ejected out of the cell.
The most downregulated protein in the MIA-GR8 cells was the S100A4, a calcium-binding protein known to promote cellular proliferation and tumor progression. The reduced expression of the protein is on par with the in vivo and in vitro studies depicting reduced cellular proliferation to escape the drug action. Furthermore, drug-resistant cells also exhibited lower counts of proteins involved in the phosphorylation of Gemcitabine into its active forms and ENT1, a transporter protein that mediates the drug entry into the cell.
Other biological processes that were altered include cell proliferation, migration, DNA repair, and apoptosis resistance. The altered protein environment induces reduced uptake and intracellular accumulation of active gemcitabine metabolites while encouraging measures to deflect the drug action on cells, such as reduced energy production and improved DNA repair mechanisms. To investigate if the mechanism of drug resistance is consistent in other models, protein quantification of PNAC-1, an intrinsically gemcitabine-resistant cell line, was performed. Though the exact adaptations adopted by PNAC-1 and MIA-GR8 differed, both focused on lowering the concentrations of active gemcitabine metabolites within the cell.
STUDY CONCLUSIONS
By developing cell lines 75x more resistant to Gemcitabine from gemcitabine-sensitive parental cell lines, the authors tried to mimic the clinical scenario of transition from treatment-receptive tumors to treatment-resistant ones. By comparing the so-developed cell lines to intrinsically resistant ones, the strong influence of Gemcitabine metabolizing pathways in imparting resistance was revealed, making them viable targets for novel therapeutic combinations. Though many of the significant proteins are marked in the study, RRM1 was highlighted as a superior candidate with strong roots in the clinical data.
Article Source: Reference Paper
Learn More:
Top Bioinformatics Books ↗
Learn more to get deeper insights into the field of bioinformatics.
Top Free Online Bioinformatics Courses ↗
Freely available courses to learn each and every aspect of bioinformatics.
Latest Bioinformatics Breakthroughs ↗
Stay updated with the latest discoveries in the field of bioinformatics.
Catherene Tomy is a consulting Content Writing Intern at the Centre of Bioinformatics Research and Technology (CBIRT). She has a master’s degree in Molecular Medicine from Amrita University with research experience in the fields of bioinformatics, cell biology, and molecular biology. She loves to pull apart complex concepts and weave a story around them.
.