Researchers from Bellvitge Biomedical Research Institute in Spain have designed TALKIEN (crossTALK IntEraction Network), a user-friendly online tool that helps to visualize molecular crosstalk or communication at the cellular level by constructing and analyzing ligand-receptor interaction network (LRIN). Understanding cellular communication is crucial for understanding cancer growth and therapy response. With the ability to perform functional analysis and access information on drugs targeting receptors, TALKIEN becomes an indispensable tool in the quest for deeper insights into cellular communication.
TALKIEN is a tool based on R (a programming language) which has immense applicability in statistical analysis and network analysis. TALKIEN takes a list of genes or proteins representing various cell types as input. It first analyzes the lists to find interactions between ligands (messengers) and receptors (receivers) on cell surfaces and then creates a map or a network of these interactions. Using systems biology techniques like centrality (measuring the importance of certain molecules), component analysis (grouping related molecules), etc., helps the users better visualize and analyze the network by providing different graphical layout options. It also shows the chain of events that occur after cell communication and presents a list of drug-targeting receptors, allowing researchers to see the broader impact of cell communication and potentially obtain unprecedented insights in healthcare.
How Minute Molecular Mechanisms Trigger Cancer
Even the smallest person can change the course of the future
The Lord of the Rings, J.R.R. – Tolkien, 1995
In the spirit of Tolkein, the interactions between the minute molecules within our cells give rise to complex processes that decide the fate of the human body. Let us take the example of cancer. Signaling molecules called cytokines, and their receptors are responsible for maintaining a critical balance in cellular communication, which prevents the uncontrollable growth of cells. Once this delicate balance is broken, cells start growing uncontrollably, giving rise to cancer. This imbalance also enables cancer cells to evade the immune system and become resistant to drugs.
Cancer is a complex disease involving communications between tumor cells themselves, the surrounding supporting cells (stromal cells), the body’s immune system, and the environment around the tumor. Hence, combating cancer not only requires the understanding of communication among cancer cells but also between cancer cells and the non-cancerous cells present around them. Many ways have been developed throughout the years to define the various kinds of cells in a tumor. Two of these methods are single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics.
Scientists may use scRNA-seq to investigate individual cells inside a tumor independently, allowing them to find changes in gene activity between tumor cells and non-cancerous cells. On the other hand, the analysis of spatial patterns of gene activity inside the tumor provides insights into the architecture of diverse cell types participating in the tumor. Proteomics is a third method that examines the protein makeup of tumor cells to reveal details on the molecular behavior of the tumor.
The methods mentioned above unveil the genetic composition, molecular composition, and characteristics of tumors. However, a technique that can unmask how affected cells interact with each other and with unaffected cells and what the underlying molecular mechanisms are, as well as the cascading after-effects, can help in treating cancer and preventing cancer.
TALKIEN: Turning Omics Data into Insights on Cell Communication in Cancer
To benefit from the observations obtained from the aforementioned technologies and understand the molecular crosstalk that takes place within a tumor, Ferran Moratalla-Navarro and his team have developed TALKIEN. TALKIEN is a web-based tool that creates networks of interactions between proteins in the cells and aids in deciphering how they help the cells communicate and what happens due to these communications in the tumor. It also suggests potential drug targets to interfere with these communications. The primary principle behind TALKIEN is that signaling-related communication in cells is associated with the physical interaction between signaling molecules secreted by some cells and specific proteins present in other cells, which ultimately influences their behavior. Hence, for understanding these communications, knowing about these interactions and the pathways they activate are important prerequisites.
The good news is that TALKIEN has already been used by researchers to analyze data from tumor samples in not one but multiple case studies. The first study analyzed the differential genetic expression between colorectal tumors, their surrounding mucosa, and between the surrounding mucosa of affected people and healthy donors. Among the crucial genes identified, ITGA2 emerged as a key player, displaying high activity in tumor tissues and engaging in interactions with several genes that were actively expressed in normal tissues. This finding pointed to the potential significance of ITGA2 in tumor biology and beyond. Regarding therapeutic possibilities, the network analysis identified drugs that could potentially inhibit ITGA2 activity. Among them, Vatelizumab, an agent tested in inflammatory diseases, emerged as a promising candidate for further investigation in the context of this molecular crosstalk.
An additional investigation into colorectal malignancies found that interactions between the tumors and cancer-associated fibroblasts (CAFs) cause a signaling cascade that renders the cells resistant to treatment. The in-vivo experiments also showed that the disruption of this interaction makes the cancer cells vulnerable to chemotherapy again. Another study attempted to investigate the communication between tumor cells and the surrounding stroma in a lung cancer patient who had undergone Erlotinib treatment. Employing TALKIEN, a “crosstalk network” was meticulously constructed, which revealed that the stroma displayed a higher number of activated receptors compared to the tumor cells, suggesting active stimulation by the tumor cells. One specific receptor, CD44, emerged as a pivotal player, engaging in numerous interactions with ligands from the tumor cells, forming a highly interconnected component within the network. Downstream pathways in the stroma, particularly related to interferon signaling, pointed to the immune system’s potential role in the patient’s cancer recurrence. Excitingly, TALKIEN’s analysis identified potential therapeutic targets, including specific drugs with the potential to inhibit these receptors, such as immunotherapy-based treatments.
Finally, a proteomics-based investigation was conducted to examine data from various immune cell types found in human blood. The study revealed that different immune cells in the blood do not actively interact with each other or establish strong communication pathways, at least under normal physiological conditions.
The Marvellous Methodology of TALKIEN
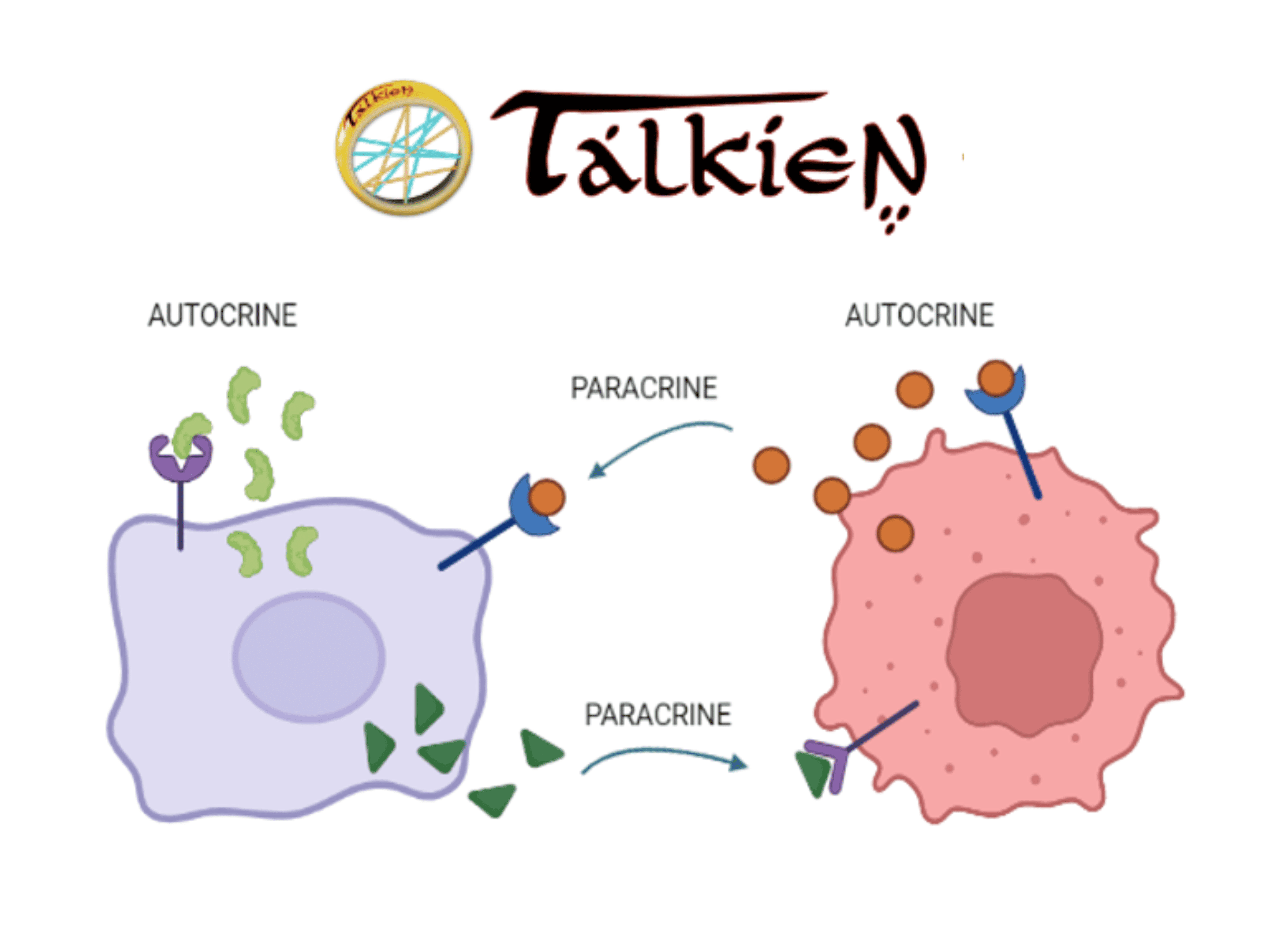
TALKIEN constructs and analyzes highly confident networks based on ligand-receptor interactions (LRI) and intracellular protein-protein interactions (PPI). It offers two types of networks for analysis: the “crosstalk network,” which includes paracrine LRIs (communication between nearby cells), and the “full network,” which encompasses both paracrine and autocrine LRIs (self-signaling).
Using TALKIEN is simple. Users provide lists of genes or proteins they wish to study, and the tool combines data from four cutting-edge LRI databases: CellPhoneDB, CellChat, iCellNet, and data extracted from Ramilowski et al. The resulting interactions are homogenized to remove incongruent annotations and create a comprehensive list of 732 ligands, 631 receptors, and 2691 interactions.
To further enrich the network with downstream intracellular interactions, TALKIEN taps into the STRING Protein-Protein Interaction Networks (PPINs). It retrieves two subnetworks—one with highly confident interactions (combined STRING score > 0.7) and another with curated interactions (combined STRING score > 0.3) —allowing users to choose their preference for intracellular interactions.
TALKIEN provides versatile network graphical layouts, ensuring optimal visualization for various networks. It offers detailed network parameters, including node-specific measures and network centrality measures, giving researchers a comprehensive view of the network’s structure and communication strength.
Functional analysis is another powerful feature of TALKIEN. Researchers can perform Reactome-based enrichment analysis on the full network or specific parts, uncovering the biological pathways and functions associated with the proteins in the network, and the results that have adjusted p-values below 0.05 are considered significant.
Not stopping there, TALKIEN empowers users to explore pathways or signals activated downstream of specific receptors of interest, providing insights into intracellular signaling events. Moreover, the tool checks for available drug-gene interactions for each receptor in the network, leveraging the DGIdb (Drug Gene Interaction Database) to identify potential drug targets.
TALKIEN vs. ITS PEERS!
TALKIEN is not the only tool that exists for inferring cell-cell communication. Many similar tools, such as Omnipath, CellChat, CytoTalk, etc., have also been developed to decode ligand-receptor interactions. However, most of them utilize gene coexpression (how genes are simultaneously active) for analyzing those interactions. In contrast, TALKIEN looks at the presence or absence of gene expression or protein abundance in different cell types to understand how cells communicate with each other. The issue with gene coexpression is that it does not always immediately transfer into the same patterns at the protein level from the coexpression of genes at the transcriptome level. Some TALKIEN data was implemented in other tools like iTALK, which generated a comparable network size, however, it failed at capturing autocrine signaling interactions.
The biggest advantage of TALKIEN is that it is highly user-friendly, allowing scientists with limited computational expertise to use it conveniently. Keeping in mind that every tool has strengths and weaknesses, it is important to highlight that TALKIEN cannot deal with raw data and analyze gene correlations. It assumes that primary analyses have already been conducted. The future scope of TALKIEN will be checked and updated every six months, and the researchers plan to release a modified version capable of dealing with mice interactions.
Conclusion
From unraveling the complexities of tumor-stroma interactions in cancer patients to shedding light on immune cell signaling in the blood, TALKIEN’s capabilities have paved the way for groundbreaking discoveries in various fields of biomedical research. Its user-friendliness is what gives it an edge over tools, as it makes network analysis in biology accessible to a wider audience. TALKIEN’s future appears bright as scientific methods are constantly evolving, and scientists even plan to upgrade TALKIEN, which can drive advancements in healthcare, improving numerous lives.
Story Source: Reference Paper | TALKIEN is freely available: Website
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.