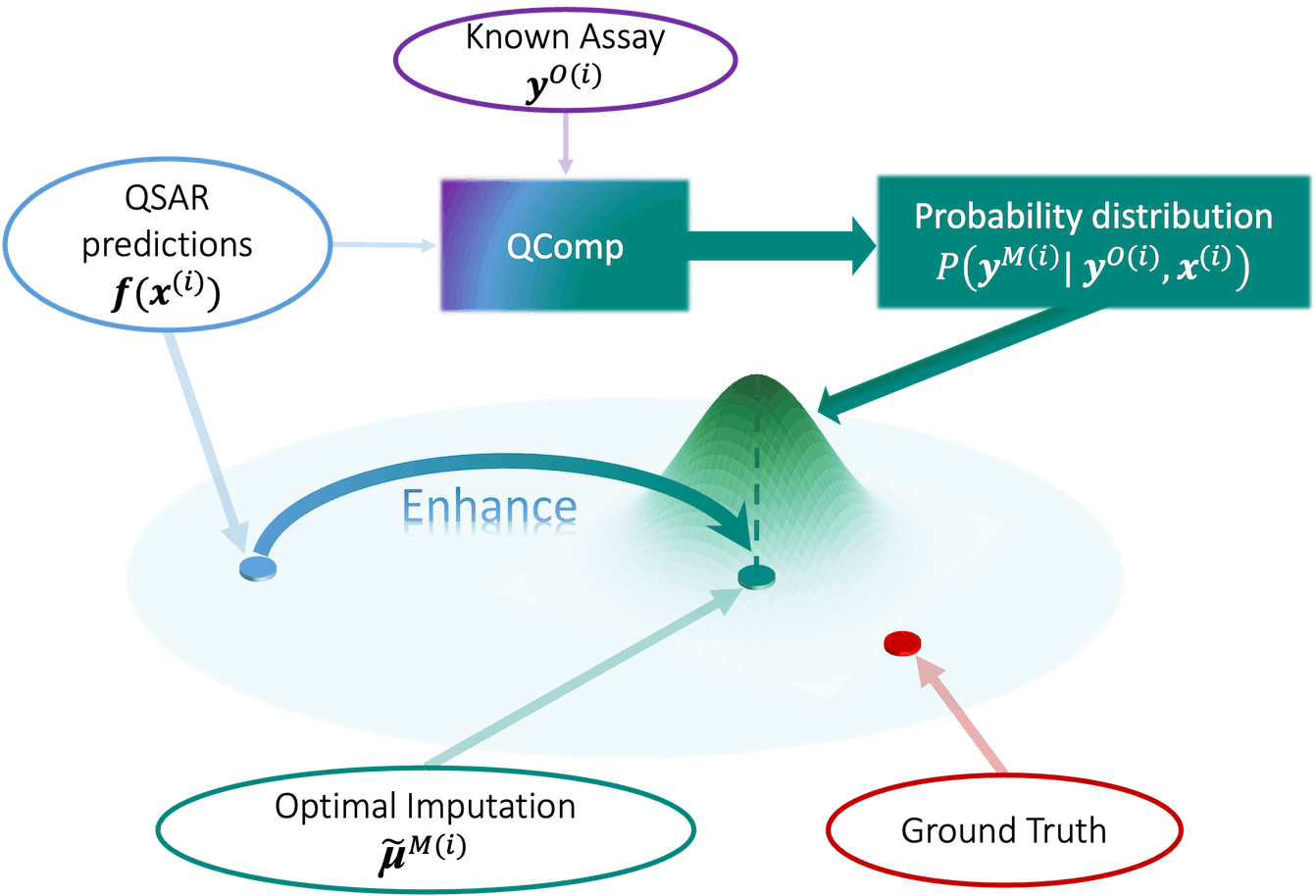
In drug discovery, the accurate prediction of chemical activities is paramount. The conventional forms of QSAR that were initially developed and widely used are the traditional or classical approach of QSAR modeling. However, there is a fundamentally new approach that redefines the future of data completion in drug discovery, called QSAR-Complete (QComp), developed by the Department of Chemistry at Princeton University in collaboration with the Department of Mathematics and Statistics at McGill University and various departments within Merck & Co., Inc.
The Rise of QComp
QComp stands out for its ability to leverage sparse data effectively, making it a game-changer in the industry. By tapping into the intrinsic correlation between chemical activities, QComp offers a unique advantage over traditional QSAR models. Its innovative approach allows for instant integration of newly acquired data, setting it apart from its predecessors.
The Need for Data Completion
Data Completion is used in the framework of drug discovery to complete records in cases where there is little data available for a compound. However, conflicts may occur when updating experimental data in existing traditional QSAR models. QComp is an effective method for filling missing values because it is built upon previously developed QSAR models, enhances predictability, and offers orientation on which experiments should be carried out to design new drugs in pharmaceutical science.
Advantages Over Traditional Models
QComp is more effective compared to other standard QSAR models and data completion techniques since it creates a higher performance, able to handle immense data sets. QComp demonstrates the progressive improvements made while increasing accuracy in the presentation. This shows how the proposed method is capable of outcompeting existing methods such as MICE, Missforest, and Macau, pointing to the fact that the newly developed method is likely to act as a marker in the field.
Experimental Validation
QComp, a recently proposed data completion method, exploits the relationship of chemical activity to complete the datasets and make effective utilization of newly obtained sparse data immediately. Compared to other QSAR techniques, QComp can incorporate new data within the model without recalibration.
Even when compared to similar work that structures ADMET data around QSAR models, QComp generates results superior to chemprop, random forest, MICE, MissForest, and Macau. Still, other times, other approaches do not work, but QComp does an excellent job of matching base QSAR performances.
In drug discovery, QComp shows significant improvements:
- Animal to Human Assay Predictions: Improves r² levels of ~ 0. 5 to > 0. 7.
- Peptide Drug Discovery: Compared to traditional methodologies for developing QSAR models, this method performs better.
- Preclinical Decision-Making: Over-in vitro basal assay prediction is favored over in vivo basal assay prediction.
Therefore, it could be considered that QComp is accurate, robust, interpretable, and versatile, integrating easily into existing QSAR workflows at low cost and showing promise for future material discovery applications. One key advantage of the technique lies in its ability to effectively build on the assay-assay correlations, which firmly establishes it as a state-of-the-art tool in the industry.
Addressing Data Completion Uncertainty
To tackle such issues concerning data completion, QComp adopted a composite uncertainty technique to give an accurate prediction. Altogether, by specifying how the problem can be avoided at best or at least mitigated, QComp provides an ultimate spectrum of data completion methodologies.
Limitations
The existing drawback at the present stage of QComp is based on the fact that it uses only one covariance matrix for all compounds of the database. Though the procedure may look slightly more complicated than before, even introducing a compound-dependent covariance matrix could add a level of detail to the probability density function. However, since the benchmarking is performed with only one instance of the base QSAR model during the QComp training, we cannot test concurrent training here. Although one of the primary benefits of QComp is it can supplement any existing QSAR model, the evaluation of a few of the integrated models like Alchemite and pQSAR, the idea of the concurrent training could improve QComp’s performance.
Further, the greedy scheme proposed for rational choice fails to capture the nuances of considerations within the higher economic and ethical costs of each experiment. It must be noted that it is possible to set up an objective function with the ability to treat gain of certainty (GOC) alongside the cost of each experiment.
Conclusion
Analyzing the significance of data completion in the context of modern tendencies in the sphere of pharmaceutical companies and brand management is very crucial these days. As QComp now takes up the mantle, the prospects for this area of compound predictive modeling and chemical activity predictions are becoming brighter. In a position to set high standards for the future of drug discovery, QComp challenges old paradigms and steps forward to a more precise future.
To gain more insights about QComp, kindly refer to the Reference Article !! QComp code is available on GitHub.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anshika is a consulting scientific writing intern at CBIRT with a strong passion for drug discovery and design. Currently pursuing a BTech in Biotechnology, she endeavors to unite her proficiency in technology with her biological aspirations. Anshika is deeply interested in structural bioinformatics and computational biology. She is committed to simplifying complex scientific concepts, ensuring they are understandable to a wide range of audiences through her writing.