BioAutoMATED, a novel, automated machine-learning tool, has eliminated the biggest barrier preventing life scientists from effectively utilizing machine learning (ML) in the analysis and design of biological sequences. This platform, developed by a group of scientists from Harvard University and MIT, brings together multiple automated machine learning (Auto ML) techniques into a unified framework, making it easier for life scientists with limited ML expertise to include machine learning in their research. It automatically provides suitable techniques for analyzing and designing biological sequences. BioAutoMATED not only analyzes protein-drug interactions, gene regulation, and glycan sequences but also aids in designing optimized synthetic biological components, proving itself to be a versatile and indispensable tool for life science researchers.
Machine Learning and Biologists: A Tricky Relationship
The development of extensive, high-dimensional biological datasets in combination with machine learning (ML) techniques has catapulted the delivery of remarkable breakthroughs in genomics and increased the potential for such breakthroughs in systems biology, synthetic biology, and structural biology. Biological sequence datasets such as nucleic acids, peptides, and glycans are widespread. Application of ML methods to such datasets can enhance the designing of sequences containing desired characteristics.
Even though life science researchers have access to ML methods through online tutorials, software packages, and open-source code, designing, training, and deploying ML models suited for processing biological sequence data without sufficient expertise proves to be an obstacle to biologists wanting to incorporate ML into their research. Not only that, but highly skilled ML practitioners also find it difficult to design models involving the selection of suitable algorithms and optimization of parameters, especially when the parameters range from thousands to hundreds of millions.
The need of the hour is a technique or tool that can eliminate the shortcomings of manual ML model designing or a tool that can automate such a process, isn’t it? Well, the need has been fulfilled through automated machine learning.
AutoML: A Revolutionary Alternative to Conventional ML
Automated Machine Learning comes to the rescue against the hurdles faced by biologists wanting to adopt machine learning in life science research. Automated machine learning includes methods that are capable of automatically designing and deploying ML pipelines with minimal manual intervention. Automated machine learning makes it convenient for biologists to pre-process data, extract features, select and optimize models and evaluate performance. It effectively determines optimized model architectures and hyperparameter values, making the process of generating initial predictive models a smooth experience for biologists. It also benefits experienced ML practitioners by allowing them to quickly build baseline models for comparison and categorization of the models.
Best Auto ML Tool for Biologists, is there any?
An immense variety of AutoML tools exist. Many established tools implement classes of neural network models, and the remaining ones use tree-based optimization methods. The tools using the tree-based method, such as random forest classifiers, are the most exciting to work with, but they have one major limitation; they are applicable to smaller and sparser biological datasets. Additionally, they have not been used in integration with neural network-based search methods yet to expedite biological sequence analysis. Moreover, recent studies emphasize the fact that there exists no “best” AutoML tool. This highlights the need for a platform that can unify multiple AutoML methods in a single framework with the ability to provide relevant techniques for data pre-processing, model deployment, and system reporting while ensuring scalability.
Now, the question arises, can such a platform be developed, or has such a platform already been developed?
BioAutoMATED: A Boon for Biologists
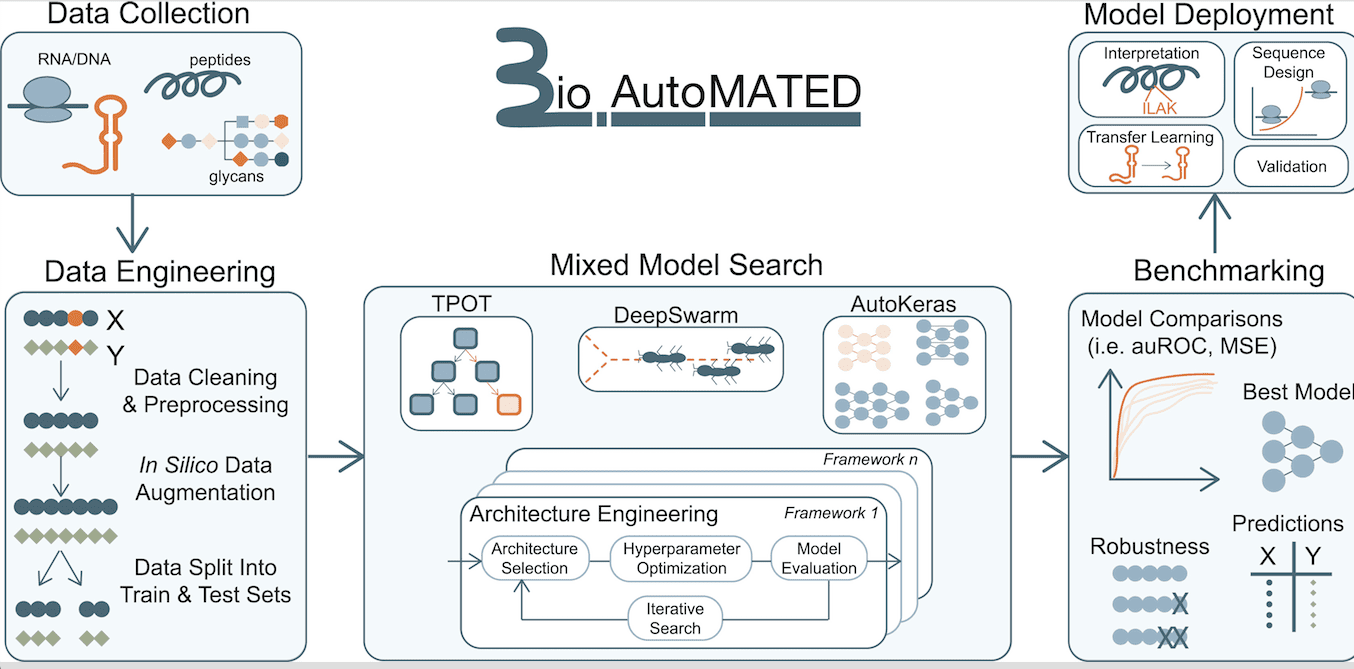
Valeri, Soenksen, Collins, et al. have developed a Python-based AutoML platform meant for building models with glycan, nucleic acid, and peptide sequence inputs. BioAutoMATED brings together multiple open-source AutoML tools with a variety of search mechanisms while also including inbuilt functionalities of data importing, pre-processing, architecture selection, hyperparameter search, model deployment, and performance reporting in an easy-to-use high-level programming interface accessible via a Jupyter Notebook. BioAutoMATED aids in the analysis and interpretation of biological models through automated prediction of crucial regions and motifs within sequences. It also enables the computer-aided designing of new and original biological sequences.
To demonstrate the utility of BioAutoMATED, researchers tested and deployed ML models based on various datasets pertaining to gene regulation, peptide-drug interaction, and glycan annotation. In each of these cases, the inputs, outputs, and features that the model learned were:
- Gene Regulation
Input: Ribosome binding site sequence in Escherichia coli
Output: Translation efficiency
Features: BioAutoMATED identified a model that could predict the translation efficiency within 30 minutes of runtime from merely ten lines of user input. Its performance was comparable to manually optimized models.
- Peptide-drug Interactions
Input: Antibody sequences varying in their CDR-H3 (Complexity Determining Region 3 of the heavy chain of an antibody) regions
Output: Binding affinity to drug ranibizumab as a target antigen
Features: BioAutoMATED produced highly predictive models capable of providing information on the development of antibody variants with improved target specificity.
- Glycan Annotation
Input: Glycan sequence
Output: Taxonomic group classification and immunogenicity in humans
Features: BioAutoMATED identified the best-performing model for the prediction of glycan immunogenicity in humans. It also identified a model that can recognize phylogenetic domains based on sequence information, thus, helping in their annotation.
Additionally, it was also applied to a study associated with RNA toehold switch design for the detection of nucleic acids, and it could identify models that could predict the performance of toehold switches capable of detecting RNA from the Zika virus.
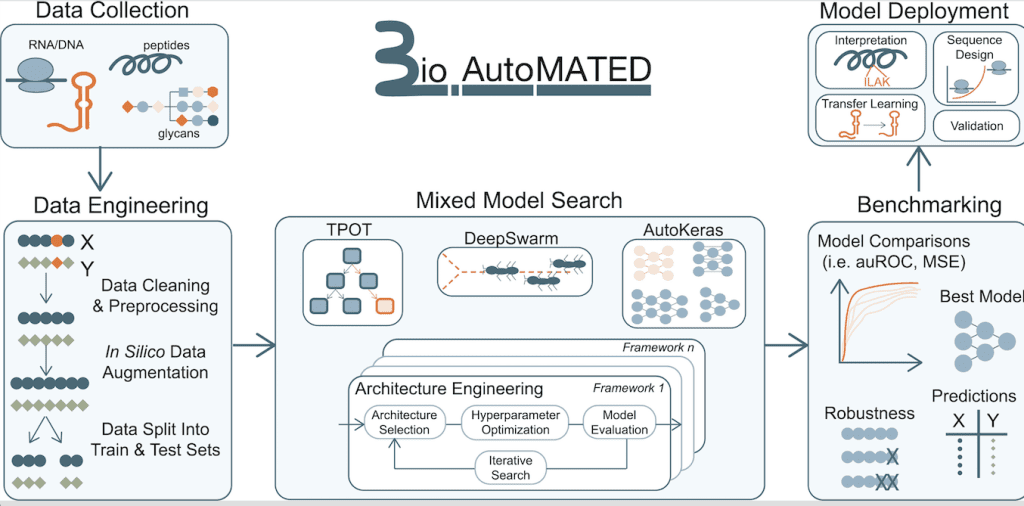
Image Source: https://doi.org/10.1016/j.cels.2023.05.007
Benefits Bestowed by BioAutoMATED to Biologists
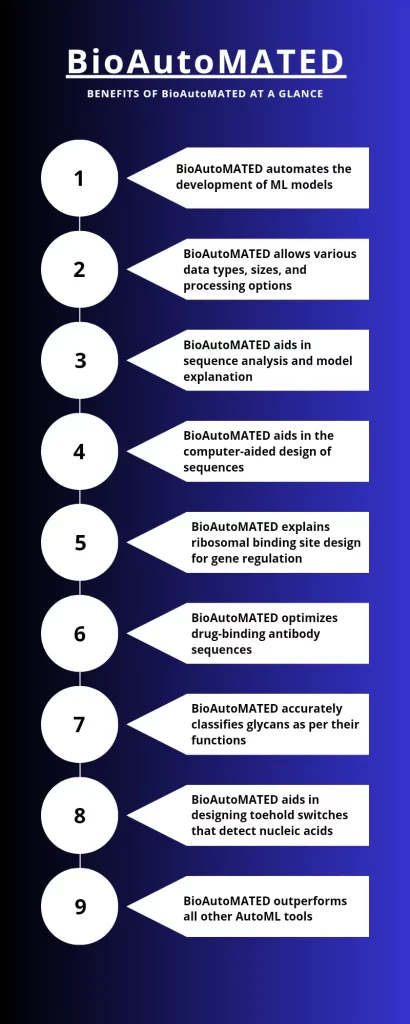
Image Credit: Neegar@CBIRT
BioAutoMATED has greatly simplified the incorporation of machine learning in biological research. It provides the convenience of automating the development of ML models suitable for handling datasets containing biological sequences, including those of nucleic acids, glycans, and peptides. The user has to upload an Excel or CSV file containing the list of biological sequences as inputs and corresponding target values that are to be predicted as outputs. BioAutoMATED then employs AutoML tools combining DeepSwarm, AutoKeras, and TPOT to perform architecture and hyperparameter searches. Finally, the most optimized model is found, and results from all three systems are evaluated within BioAutoMATED.
BioAutoMATED expands the horizon of user involvement and convenience by accommodating various data types, sizes, and processing options. It also provides tools for sequential analysis as well as model explanation. It also facilitates the computer-aided designing of novel biological sequences. Moreover, it outperformed other AutoML tools, including iLearnPlus, BioAutoML, and JADBio.
Conclusion
Life science is a vast ocean. Integrating it with other disciplines, such as computer science and mathematics, can accelerate the research process and provide valuable insights that could potentially solve humanity’s biggest crises. Thus, biologists must be empowered with interdisciplinary knowledge and powerful tools backed by technologies such as machine learning. Until recently, the greatest barrier to the utilization of machine learning by biologists was the immense experience required to build, train, and deploy models effectively. However, the advent of AutoML and the integration of various powerful AutoML tools into platforms such as BioAutoMATED has expanded the scope for biologists to conveniently incorporate ML into their research, which can rapidly fuel novel discoveries and inventions in all the various domains of biology. BioAutoMATED provides a plethora of benefits to biologists, along with outperforming the various other AutoML tools. This opens the door to exciting new prospects and areas for pioneering study and development in the field of biology.
Article Source: Reference Paper
Learn More:
Neegar is a consulting scientific content writing intern at CBIRT. She's a final-year student pursuing a B.Tech in Biotechnology at Odisha University of Technology and Research. Neegar's enthusiasm is sparked by the dynamic and interdisciplinary aspects of bioinformatics. She possesses a remarkable ability to elucidate intricate concepts using accessible language. Consequently, she aspires to amalgamate her proficiency in bioinformatics with her passion for writing, aiming to convey pioneering breakthroughs and innovations in the field of bioinformatics in a comprehensible manner to a wide audience.